CN103314003A - Compositions and methods for activating expression by specific endogenous miRNA - Google Patents
Compositions and methods for activating expression by specific endogenous miRNA Download PDFInfo
- Publication number
- CN103314003A CN103314003A CN2011800631535A CN201180063153A CN103314003A CN 103314003 A CN103314003 A CN 103314003A CN 2011800631535 A CN2011800631535 A CN 2011800631535A CN 201180063153 A CN201180063153 A CN 201180063153A CN 103314003 A CN103314003 A CN 103314003A
- Authority
- CN
- China
- Prior art keywords
- mir
- sequence
- exogenous
- composition
- rna molecule
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Pending
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07H—SUGARS; DERIVATIVES THEREOF; NUCLEOSIDES; NUCLEOTIDES; NUCLEIC ACIDS
- C07H21/00—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids
- C07H21/02—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids with ribosyl as saccharide radical
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K31/00—Medicinal preparations containing organic active ingredients
- A61K31/70—Carbohydrates; Sugars; Derivatives thereof
- A61K31/7088—Compounds having three or more nucleosides or nucleotides
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K48/00—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy
- A61K48/005—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'active' part of the composition delivered, i.e. the nucleic acid delivered
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K48/00—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy
- A61K48/005—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'active' part of the composition delivered, i.e. the nucleic acid delivered
- A61K48/0066—Manipulation of the nucleic acid to modify its expression pattern, e.g. enhance its duration of expression, achieved by the presence of particular introns in the delivered nucleic acid
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P43/00—Drugs for specific purposes, not provided for in groups A61P1/00-A61P41/00
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07H—SUGARS; DERIVATIVES THEREOF; NUCLEOSIDES; NUCLEOTIDES; NUCLEIC ACIDS
- C07H21/00—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids
- C07H21/04—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids with deoxyribosyl as saccharide radical
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/635—Externally inducible repressor mediated regulation of gene expression, e.g. tetR inducible by tetracyline
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/111—General methods applicable to biologically active non-coding nucleic acids
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/10—Type of nucleic acid
- C12N2310/14—Type of nucleic acid interfering N.A.
- C12N2310/141—MicroRNAs, miRNAs
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2320/00—Applications; Uses
-
- Y—GENERAL TAGGING OF NEW TECHNOLOGICAL DEVELOPMENTS; GENERAL TAGGING OF CROSS-SECTIONAL TECHNOLOGIES SPANNING OVER SEVERAL SECTIONS OF THE IPC; TECHNICAL SUBJECTS COVERED BY FORMER USPC CROSS-REFERENCE ART COLLECTIONS [XRACs] AND DIGESTS
- Y02—TECHNOLOGIES OR APPLICATIONS FOR MITIGATION OR ADAPTATION AGAINST CLIMATE CHANGE
- Y02A—TECHNOLOGIES FOR ADAPTATION TO CLIMATE CHANGE
- Y02A50/00—TECHNOLOGIES FOR ADAPTATION TO CLIMATE CHANGE in human health protection, e.g. against extreme weather
- Y02A50/30—Against vector-borne diseases, e.g. mosquito-borne, fly-borne, tick-borne or waterborne diseases whose impact is exacerbated by climate change
Abstract
There are provided compositions and methods for activating expression of an exogenous polynucleotide of interest only in the presence of a specific endogenous miRNA in a cell. Further provided are uses for the compositions in treatment and diagnosis of various conditions and disorders, for example by selectively activating expression of a toxin only in target cell populations.
Description
Invention field
The present invention relates to activate when only in cell, having miRNAs in the specificity composition that the external source polynucleotide of interest is expressed.As by only in the target cell group selectively activate toxin express illustratedly, the invention still further relates to these compositions in treatment and diagnose purposes in various illnesss and the illness.
Background of invention
Virus is the second important risk factors that biological entities type the abundantest on the earth and virus are rendered as the human cancer development.WHO (World Health Organization) international cancer research office estimates that in 2002 ,~15% human cancer was to be caused by 7 kinds of different viruses.Because the oncogene in its genome, virus may be carcinogenic.Because with gene brachymemma or the integration that gene placed the site under the control of strong viral cis acting controlling element, retrovirus also may be carcinogenic.According to WHO, in 2006, there was the people of about 3,950 ten thousand trouble HIV in the whole world.The many viruses that comprise HIV show resting stage or latent period, carry out few protein synthesis or do not carry out protein synthesis during described resting stage or latent period.During these stages, virus infection is sightless for immunity system basically.Present antiviral therapy scheme is invalid [1] in major part aspect the cell reserves of eliminating latent virus.
According to American Cancer Society, during 2007,7,600,000 people die from cancer in the world.Every kind of tumour comprises average 90 kinds of mutator genes [2], and wherein every kind of tumour originates from independent founder cell [33].Character and the basic skills of cancer therapy constantly change.For example the method for radiotherapy, operation and the angiopoietic cancer therapy of inhibition is useless for many little metastasis.The method that for example suppresses the cancer therapy of the cell in cell fission and the destruction division does not have specificity and therefore produces the harmful side effect that the patient is caused death.For example the induced tumor tissue differentiation, suppress oncogene, comprise virus at the part of the peculiar membrane receptor protein of cancer cells, handle the method for the cancer therapy of immunity system and immunotoxin treatment, have narrow therapeutic index and usually enough not effectively.Use tumor suppressor gene cancer therapy method and use the narrow therapeutic index of the method tool of cancer therapy of the toxin under the promotor that in cancer cells, is activated uniquely, produce the big possibility of harmful side effect and usually enough not effectively.
Ribosome inactivating protein (RIP) is plant or microbe-derived archon.RIP is by making rrna inactivation arrestin matter synthetic.The nearest RIP that studies show that can also pass through the apoptosis induction necrocytosis.II type RIP comprises toxin A chain and the agglutinin subunit (B chain) that links together by disulfide linkage.B chain non-activity in catalysis enters cytosol but be used for regulating the A-B albumen composition.Ricin, abrin and diphtheria toxin are very potent II type RIP.Reported that the unit molecule Ricin or the abrin that arrive cytosol can kill this cell [3,4].In addition, the unit molecule diphtheria toxin Segment A that is incorporated in the cell can be killed this cell [5].
In mammalian cell, add cap (7-methylguanosine cap) to 5 of mRNA ' end and make the mRNA translation be increased to 35-50 doubly.In addition, adding poly (A) tail to 3 of mRNA ' end makes the mRNA translation be increased to 114-155 doubly [6].Poly in the mammalian cell (A) tail only makes the transformation period of functional mRNA be increased to 2.6 times and cap only makes the transformation period of functional mRNA be increased to 1.7 times [6].A member of people HIST1H2AC (H2ac) genes encoding histone H2A family.Lack poly (A) tail from this gene transcription thing, but alternatively comprise palindrome termination element (5 '-GGCUCUUUUCAGAGCC-3 '), this element 3 '-the UTR place be formed on mRNA processing and stable aspect conservative stem-ring structure [7] of playing an important role.
It is wherein by treating that with function the dsRNA that has adopted RNA and sense-rna to constitute of the specific region homology of repressed target gene influences a kind of phenomenon of cracking in homology zone of the transcript of described target gene that RNA disturbs (RNAi).In Mammals, dsRNA should be shorter than 31 base pairs to avoid causing the ifn response that can cause necrocytosis by apoptosis.Medical science in 2006 and the physiology Nobel prize have been authorized RNAi the field, because this technology has huge treatment potentiality.Yet the RNAi technology is based on utilizing Microrna (miRNA) to regulate the natural mechanism [8] that posttranscriptional gene is expressed.MiRNA is the very little RNA molecule of about 21 Nucleotide of length, and it shows the precursor derived from 70-90 the Nucleotide (nt) of the RNA stem-ring structure that forms prediction.MiRNA for example expresses in nematode, fruit bat, people and the plant different organisms.
In Mammals, miRNA transcribed by rna plymerase ii usually and the primary transcript of gained (elementary-as miRNA) to comprise by the local stem-ring structure of Drosha-DGCR8 mixture cracking.The product of this cracking be one or more (under the situation of bunch collection) precursor miRNA (preceding-miRNA).Before-the miRNA normal length is 70-90 Nucleotide, has firm stem-ring structure, and it comprises the overhang [9] of 2 Nucleotide usually at 3 ' end.Preceding-miRNA is output albumen-5 and is transported in the tenuigenin.In tenuigenin, the stem among preceding-miRNA is identified as the dsRNA (miRNA duplex) of 3 of dsRNA and the past-miRNA ' and 5 ' end check solution and release 21bp for the Dicer enzyme of the endoribonuclease of RNA enzyme III family.Dicer-TRBP mixture chain two chains of duplex are separated from one another and that have more weak 5 ' end on the thermokinetics is incorporated in the reticent mixture of RNA inducibility (RISC) [10].This chain is ripe miRNA.The chain that is not incorporated among the RISC is called the miRNA* chain and its be degraded [8].Ripe miRNA is directed to target site in the mRNA with RISC.If target site and ripe miRNA are almost completely complementary, then this mRNA will be in the position cleaved [10] that is positioned at target site 3 ' about 10 Nucleotide in end upstream.After cracking, RISC-is ripe, and the miRNA chain cpd is recovered for another activity [11].If target site and ripe miRNA have lower complementarity, then mRNA will be not in the cracking of target site place but the translation of this mRNA will be suppressed.Though in the mankind, identified about 530 kinds of miRNA so far, estimate the nearly miRNA of 1,000 kind of uniqueness of vertebrate genome encoding, predict that these miRNA regulate at least 30% expression of gene [12], and Fig. 1.
Microrna human cancer start and progress aspect as if play crucial effects, and the Microrna that works in cancer is named as carinogenicity miRNA (oncomiR) [12].In the lung cancer that is one of cancer the most general among the grownup of developed country, the expression of miRNA bunch of miR-17-92 is raised strongly; The prediction target of miR-17-92 is PTEN and RB2, two kinds of known tumor suppressor genes [8].In papillary thyroid carcinoma (PTC), three kinds of miRNA:miR-221, miR-222 and miR-146 are with than level accumulation [8] much higher in the health tissues of coupling.In the cancer of the brain glioblastoma multiforme (GBM) of most common form, miR-221 and miR-21 are to accumulate [8] than level much higher in healthy tissues.In the lymphoma in lymphocyte cancer B-cell source, miR-155 is to accumulate [8] than level much higher in normal lymphocyte.Compare with health or non-metastatic tumorigenic cell, in metastatic breast cancer, transcription factor Twist raises miR-10b and expresses; The target of miR-10b is HOXD10, and the level of reduction HOXD10 causes higher levels of RHOC, the mobility [8] of RHOC irritation cancer cell.
The full genome screening that becomes feasible because of the checking of computer approach and high-throughput has had been found that about 141 kinds of Microrna precursors [34 by encoding viral, 35], the major part in these Micrornas is by the simplexvirus family coding [13] that comprises various human oncovirus such as hsv, Kaposi sarcoma simplexvirus or Epstein epstein-Barr virus.Many viral miRNA be arranged in latent transcribe relevant genome area or should the zone around bunch [20].Shown three kinds of herpes simplex virus group hsvs-1 (HSV-1) and Marek's disease virus-1 and 2 (MDV-1 and MDV-2) coding near miRNA and the miRNA in described transcript of relevant transcript (the minor latency-associated transcript) that hide on a small quantity, a kind of during the latent infection of all three kinds of viruses detected non-coding RNA [20].In two genome areas of γ-simplexvirus Epstein epstein-Barr virus, identify multiple miRNA and their expression [20] during the latent infection of the B clone that transforms.In mouse γ-simplexvirus-68 (MHV-68), the tRNA-sample transcript that is accredited as the sign in latent period before the finding multiple miRNA that encodes, and be from also expressing [20] that relevant single transcript processing obtains with latent gene by most of miRNA that the relevant simplexvirus (KSHV) of Kaposi sarcoma is expressed.Other Micrornas that studies show that the HIV-coding are in influence and/or keep effect [1,14] in the latent infection.
Cause many encoding viral miRNA of cancer and can cause latent infection.For example, KSHV virus causes the Kaposi sarcoma cancer and encodes 13 kinds of miRNA[13].For example, SV40 (simian vacuolating virus 40) has the potentiality that cause tumour, but the most often continue to exist with latent infection, SV40 regulates the expression of its large T antigen by direct coding with two kinds of miRNA of gene antisense, and the expression of these miRNA causes the cracking [20] of large T antigen transcript.For example, 23 kinds of miRNA of EBV coding and at B cell Burkitt lymphoma, infected the expression [13,21] of observing EBV miRNA in the nasopharyngeal carcinoma cell of the EBV cancer of the stomach (EBVaGC) relevant with EBV-.For example, 15 kinds of miRNA of HCMV coding and the nearest genome that studies show that HCMV and antigen is greater than 90% the specific malignant tumour of trouble, for example the existing of the patient's of colorectal carcinoma, glioblastoma, prostate cancer and mammary cancer tumour cell (rather than in adjacent healthy tissues) [36].In addition, detect HCMV and show in different tissues is learned the glioma of type, compare with 48% in the tumour of even lower level, the HCMV positive cell in the glioblastoma multiforme is 79%[36].HCMV can increase the pernicious of tumour cell, because the total many interests of these tumour cells (for example, Nucleotide is synthetic, dna replication dna, escape immunity system and escape apoptosis).Present antiviral therapy scheme is invalid in major part aspect the cell reserves of eliminating latent virus.
Some viral miRNA be oncomiR (miRNA of known participation cancer) directly to homologue (the similar gene each other in the different plant species is because it is derived from common ancestors) [35].Directly to the example of the viral miRNA of homology be for hsa-miR-155 directly to the KSHV-miR-K12-11 of the KSHV of homologue, its: cross in B cell lymphoma, leukemia, carcinoma of the pancreas and the mammary cancer and express [35].Another example be for hsa-miR-18a/b directly to the EBV-miR-BART5 of the EBV of homologue.Hsa-miR-18a/b is by at the hsa-miR-17-92 bunch of coding [35] of cross expressing in lung cancer, undifferentiated thyroid carcinoma cell and the human B cell lymphoma.
Herpes virus hominis 6 (HHV6) has been accredited as cause of disease material possible in multiple sclerosis, myocarditis, encephalitis and the febrile convulsion.The sign [37] that active HHV6B copies in the patient's of about 2/3rds trouble MTS (medial temporal lobe sclerosis) hippocampal astrocytes that studies show that to temporal lobe excision art sample.HHV6 is the member of β herpetoviridae (subfamily of herpetoviridae), the β herpetoviridae also comprise HCMV (it comprises 15 kinds of miRNA) and therefore HHV6 also can comprise many miRNA.
The multiple treatment potentiality of miRNA have been proposed.A kind of method is logically to make up at the very Microrna of conservative region or short hairpin RNA (shRNA) [8] in the virus transcription thing or in the oncogene transcript of target cell; Yet in the method, the cracking of virus transcription thing or oncogene transcript can not killed target cell usually.Additive method is by anti--miRNA oligonucleotide (AMO) blocking-up carinogenicity miRNA or viral miRNA.AMO has with the sequence of miRNA complementation and comprises the chemically modified that realizes strong combination, the described miRNA of described chemically modified titratable (titrate away), one type modification be 2 of RNA Nucleotide '-O-methylates and the modification of other types is lock nucleic acid (LNA) DNA Nucleotide [8].Yet, this method has at least two problems: first, the blocking-up of the oncogene of AMO or viral miRNA can not killed target cell usually, and the second, AMO can not transit cell record and therefore AMO need be inserted into each target cell with the enormous amount that is used for the most of miRNA copy number of titration.For example, relate to the purposes that the genophore that comprises miRNA sequence target and prevention or minimizing thereof comprise the transit cell expression of gene of corresponding miRNA as disclosed another kind of method among the WO07/00068.For example, also disclose among the WO2010/055413 and be suitable for the genetically modified genophore that is operatively connected genetically modified regulating and controlling sequence that comprises of temporary transient expression in the periphery organ cell, genetically modified expression described in wherein said regulating and controlling sequence prevention or the minimizing vascularization pedigree cell.
Therefore exist potently, reliable, special and use safety and can optionally express and/or activate the particular target cell that only comprises miRNAs in the specificity rather than the needs of the novel compositions of the exogenous object protein in any other cell for development, said composition does not comprise miRNAs in the described specificity.These compositions should preferably can optionally kill the target cell that comprises miRNAs in the described specificity, and to other cells of not comprising miRNAs in the described specificity without any effect.
Summary of the invention
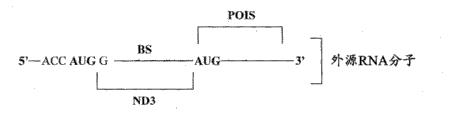
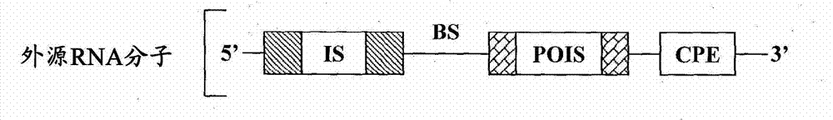
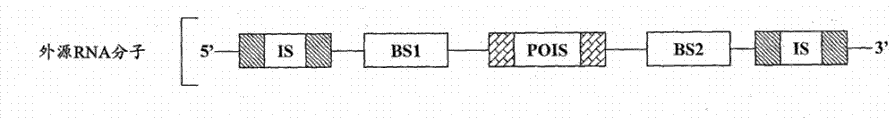
According to some embodiment, provide the composition that is used for the existence of response cell specificity endogenous cell miRNA or viral miRNA and expresses exogenous object protein.These compositions comprise or transcribe the exogenous RNA molecule, and described exogenous RNA molecule is to comprise following RNA molecule:
(a) sequence of encoding exogenous target protein;
(b) can suppress the inhibition sequence that described exogenous object protein is expressed; With
(c) the ripe miRNA chain with the interior miRNAs of described specificity has the described exogenous RNA molecule of guidance at the binding site of enough complementarity of cracking site place cracking.Predetermined target cracking site is designed between the sequence that suppresses sequence and this exogenous object protein of encoding.
Therefore, when in cell, having in the specificity miRNAs, the exogenous RNA molecule this cracking site place by described specificity in miRNAs cracking and suppress sequence and separate with the sequence of encoding exogenous target protein, make exogenous object protein to be expressed.
In certain embodiments, exogenous object protein can be selected from, but is not limited to: archon Ricin, abrin and diphtheria toxin comprise fusion rotein and the albuminoid of archon.The optional any miRNA that in cell, expresses of miRNAs in the described specificity, such as, such as but not limited to cell miRNA, carinogenicity miRNA, viral miRNA and similar miRNA or its any combination.Suppress downstream or upstream that sequence can be positioned at cracking site.
According to some embodiment, the inhibition sequence that is positioned at the cracking site upstream for example can be, but be not limited to a plurality of initiator codons (initiation codon), wherein each initiator codon can be positioned at Kozak consensus sequence (or any other translation initiation element) and wherein the sequence of each initiator codon and encoding exogenous target protein not at same reading frame.In this case, these initiator codons suppress expression of exogenous object protein.In another embodiment of the present invention, the inhibition sequence that is positioned at the cracking site upstream can be such as, but not limited to the recognition site of: sorting signals, the RNA signal for locating that is used for Subcellular Localization, ubiquitin degraded signal, the element (ARE) that is rich in AU, translation repressor (translation repressor), be enough to hinder secondary structure and similar sequence or its combination of rrna scanning.In an exemplary, the exogenous RNA molecule comprises first sequence at the inhibition sequence area place of next-door neighbour's cracking site located upstream, and wherein said first sequence can be in conjunction with second sequence of next-door neighbour's cracking site downstream location.Therefore, in complete exogenous RNA molecule, first and second sequences form the secondary structure that can hinder rrna scanning, and especially, in cleaved exogenous RNA molecule, second sequence can form internal ribosome entry site (IRES) structure.
According to other embodiments, the sequence with exogenous RNA molecule of the inhibition sequence that is positioned at the cracking site upstream also can comprise sequence or the component that can be implemented in the described exogenous RNA molecule of position cracking that suppresses the sequence upstream directly or indirectly.Therefore this can reduce the translation efficiency of complete exogenous RNA molecule.
According to other embodiments, composition of the present invention also can comprise can be increased in 5 ' one or more other structures of holding the translation efficiency of exogenous RNA molecule that can be cleaved.Described one or more other structures can be such as, but not limited to: can make described exogenous RNA molecule form the nucleotide sequence of ring (circularization), therefore described nucleotide sequence can increase the translation efficiency of cleaved exogenous RNA molecule.
According to some embodiment, composition of the present invention can be used in various application, method and the technology, such as, such as, but not limited to regulate gene expression, treat various illnesss and illness, comprise various illnesss and illness such as, the various diseases diagnostics of healthy relevant illness for example, the formation of transgenic organism is used for the treatment of hyperplasia illness such as, the suicide gene therapy of cancer for example; Be used for the treatment of: inherited disease, transmissible disease be the suicide gene therapy of HIV and similar disease for example.
According to some embodiment, provide comprise for only in expression specificity the cell of miRNAs instruct the composition of one or more polynucleotide that exogenous object protein expresses, described one or more polynucleotide encoding exogenous RNA molecules, described exogenous RNA molecule comprises: the sequence of encoding exogenous target protein; The inhibition sequence that can suppress described expression of exogenous object protein; With the binding site at miRNAs in the described specificity, wherein only in described specificity miRNAs in the presence of, described exogenous RNA molecule is cleaved at the cracking site place, suppresses sequence thereby discharge from the sequence of encoding exogenous target protein, and described exogenous object protein is so as to being expressed.In certain embodiments, enough complementarity is at least 30% complementarity.In other embodiments, enough complementarity is at least 90% complementarity.
According to some embodiment, described cracking site in described binding site and described cracking site between the sequence of described inhibition sequence and the described exogenous object protein of encoding.
In certain embodiments, have at the sequence in the miRNAs in the binding site of miRNAs in the specificity and the described specificity and make that miRNAs instructs described exogenous RNA molecule in the cleaved enough complementarity of described cracking site in the described specificity.
According to other embodiments, miRNAs is the Microrna of cell, viral Microrna or the two in the specificity.In certain embodiments, the Microrna of cell is only expressed in neoplastic cell.In certain embodiments, viral Microrna is by the expressing viral that is selected from by the following group of forming: double-stranded DNA virus, single-stranded DNA viruses, diplornavirus, diplornavirus, strand (normal chain) virus, strand (minus strand) virus and retrovirus.
According to some embodiment, described exogenous object protein is toxin.The group of the following composition of the optional freedom of described toxin: Ricin, ricin A chain, abrin, abrin A chain, diphtheria toxin A chain and modified forms thereof.In certain embodiments, described toxin is selected from the group of being made up of following: alpha toxin, saporin, Zea mays RIP (maize RIP), barley RIP, wheat RIP, corn RIP (corn RIP), rye RIP, flax RIP, shiga toxin, will are congratulated sample RIP, momordin, thymidine kinase, Pokeweed antiviral protein, are spent more white tree toxalbumin, Pseudomonas exotoxin, ETA, coli cytosine deaminase and modified forms thereof.
In some other embodiments, suppress sequence and can be positioned at the upstream of cracking site and suppress sequence and can reduce translation efficiency from the exogenous object protein of exogenous RNA molecule directly or indirectly.
In certain embodiments, suppress sequence and comprise a plurality of initiator codons.In other embodiments, the sequence of each initiator codon and encoding exogenous target protein is not in same reading frame.In certain embodiments, each described initiator codon mainly by 5 '-AUG-3 ' forms.In certain embodiments, each initiator codon can be arranged in the Kozak consensus sequence.
According to other embodiments, suppressing sequence can be in conjunction with polypeptide, and wherein said polypeptide can reduce the translation efficiency of exogenous object protein described in the described exogenous RNA molecule directly or indirectly.Described polypeptide can be translation repression albumen (translation repressor protein), and wherein said translation repression albumen is endogenous translation repression albumen or by one or more polynucleotide encodings of described composition.
In certain embodiments, the inhibition sequence comprises RNA signal for locating or the endogenous miRNA binding site for Subcellular Localization.
According to some embodiment, one or more polynucleotide of described composition also can comprise the polynucleotide sequence of functional r NA that coding can suppress the expression of endogenous exonuclease directly or indirectly.
In certain embodiments, be that a plurality of binding sites and wherein said cracking site at identical or different endogenous miRNA is a plurality of cracking sites at the binding site of miRNAs in the specificity.
In certain embodiments, miRNAs is selected from the group of being made up of following: hsv1-miR-H1 in the specificity, hsv1-miR-H2, hsv1-miR-H3, hsv1-miR-H4, hsv1-miR-H5, hsv1-miR-H6, hsv2-miR-I, hcmv-miR-UL22A, hcmv-miR-UL36, hcmv-miR-UL70, hcmv-miR-UL112, hcmv-miR-UL148D, hcmv-miR-US4, hcmv-miR-US5-1, hcmv-miR-US5-2, hcmv-miR-US25-1, hcmv-miR-US25-2, hcmv-miR-US33, kshv-miR-K12-1, kshv-miR-K12-2, kshv-miR-K12-3, kshv-miR-K12-4, kshv-miR-K12-5, kshv-miR-K12-6, kshv-miR-K12-7, kshv-miR-K12-8, kshv-miR-K12-9, kshv-miR-K12-10a, kshv-miR-K12-10b, kshv-miR-K12-11, kshv-miR-K12-12, ebv-miR-BART1, ebv-miR-BART2, ebv-miR-BART3, ebv-miR-BART4, ebv-miR-BART5, ebv-miR-BART6, ebv-miR-BART7, ebv-miR-BART8, ebv-miR-BART9, ebv-miR-BART10, ebv-miR-BART11, ebv-miR-BART12, ebv-miR-BART13, ebv-miR-BART14, ebv-miR-BART15, ebv-miR-BART16, ebv-miR-BART17, ebv-miR-BART18, ebv-miR-BART19, ebv-miR-BART20, ebv-miR-BHRF1-1, ebv-miR-BHRF1-2, ebv-miR-BHRF1-3, bkv-miR-B1, jcv-miR-J1, hiv1-miR-H1, hiv1-miR-N367, hiv1-miR-TAR, sv40-miR-S1, MCPyV-miR-M1, hsv1-miR-LAT, hsv1-miR-LAT-ICP34.5, hsv2-miR-II, hsv2-miR-III, hcmv-miR-UL23, hcmv-miR-UL36-1, hcmv-miR-UL54-1, hcmv-miR-UL70-1, hcmv-miR-UL22A-1, hcmv-miR-UL112-1, hcmv-miR-UL148D-1, hcmv-miR-US4-1, hcmv-miR-US24, hcmv-miR-US33-1, hcmv-RNA β 2.7, ebv-miR-BART1-1, ebv-miR-BART1-2, ebv-miR-BART1-3, ebv-miR-BHFR1, ebv-miR-BHFR2, ebv-miR-BHFR3, hiv1-miR-TAR-5p, hiv1-miR-TAR-p, hiv1-HAAmiRNA, hiv1-VmiRNA1, hiv1-VmiRNA2, hiv1-VmiRNA3, hiv1-VmiRNA4, mir-675, hiv1-VmiRNA5, hiv2-miR-TAR2-5p, hiv2-miR-TAR2-3p, mdv1-miR-M1, mdv1-miR-M2, mdv1-miR-M3, mdv1-miR-M4, mdv1-miR-M5, mdv1-miR-M6, mdv1-miR-M7, mdv1-miR-M8, mdv1-miR-M9, mdv1-miR-M10, mdv1-miR-M11, mdv1-miR-M12, mdv1-miR-M13, mdv2-miR-M14, mdv2-miR-M15, mdv2-miR-M16, mdv2-miR-M17, mdv2-miR-M18, mdv2-miR-M19, mdv2-miR-M20, mdv2-miR-M21, mdv2-miR-M22, mdv2-miR-M23, mdv2-miR-M24, mdv2-miR-M25, mdv2-miR-M26, mdv2-miR-M27, mdv2-miR-M28, mdv2-miR-M29, mdv2-miR-M30, mcmv-miR-M23-1, mcmv-miR-M23-2, mcmv-miR-M44-1, mcmv-miR-M55-1, mcmv-miR-M87-1, mcmv-miR-M95-1, mcmv-miR-m01-1, mcmv-miR-m01-2, mcmv-miR-m01-3, mcmv-miR-m01-4, mcmv-miR-m21-1, mcmv-miR-m22-1, mcmv-miR-m59-1, mcmv-miR-m59-2, mcmv-miR-m88-1, mcmv-miR-m107-1, mcmv-miR-m108-1, mcmv-miR-m108-2, rlcv-miR-rL1-1, rlcv-miR-rL1-2, rlcv-miR-rL1-3, rlcv-miR-rL1-4, rlcv-miR-rL1-5, rlcv-miR-rL1-6, rlcv-miR-rL1-7, rlcv-miR-rL1-8, rlcv-miR-rL1-9, rlcv-miR-rL1-10, rlcv-miR-rL1-11, rlcv-miR-rL1-12, rlcv-miR-rL1-13, rlcv-miR-rL1-14, rlcv-miR-rL1-15, rlcv-miR-rL1-16, rrv-miR-rR1-1, rrv-miR-rR1-2, rrv-miR-rR1-3, rrv-miR-rR1-4, rrv-miR-rR1-5, rrv-miR-rR1-6, rrv-miR-rR1-7, mghv-miR-M1-1, mghv-miR-M1-2, mghv-miR-M1-3, mghv-miR-M1-4, mghv-miR-M1-5, mghv-miR-M1-6, mghv-miR-M1-7, mghv-miR-M1-8, mghv-miR-M1-9 and sv40-miR-S1.Its name and sequence such as database http://www.mirbase.org/. are defined.
In certain embodiments, the exogenous RNA molecule also comprises the terminator codon between (start codon) of starting codon of the described sequence that is positioned at initiator codon and coding target protein, wherein said terminator codon and described initiator codon are in the same reading frame and wherein said terminator codon is selected from the group of being made up of following: 5 '-UAA-3 ', 5 '-UAG-3 ' and 5 '-UGA-3 '.
In other embodiments, suppress the sequence upstream that sequence is positioned at the encoding exogenous target protein, wherein said inhibition sequence can form to have and be lower than-secondary structure of the folding free energy of 30kcal/mol, and wherein said secondary structure is enough to hinder the scanning rrna and arrives starting codon of described exogenous object protein.
In other embodiments, one or more polynucleotide of described composition comprise one or more dna moleculars, one or more RNA molecules or its combination.
In other embodiments, cell is selected from the group of being made up of following: people's cell, zooblast, cultured cells and vegetable cell.In certain embodiments, described cell is neoplastic cell.In other embodiments, described cell is present in the organism.
In certain embodiments, described composition is introduced in the cell.Described cell can be that neoplastic cell and its can be present in the organism.
In certain embodiments, also provide the diagnostic kit that comprises described composition.
In other embodiments, the pharmaceutical composition that comprises described composition is provided, described pharmaceutical composition comprises described one or more polynucleotide, and one or more vehicle.
In other embodiments, provide to be used for the method that target kills the target cell that comprises the interior miRNAs of specificity, described method comprises in described target cell introduces the composition that comprises described one or more polynucleotide.
According to some embodiment, the carrier of the polynucleotide sequence that comprises encoding exogenous RNA molecule is provided, wherein said exogenous RNA molecule comprises the sequence of encoding exogenous target protein; The inhibition sequence that can suppress described expression of exogenous object protein; And at the binding site of miRNAs in the specificity.Described carrier can be virus vector.Described carrier can be non-virus carrier.In certain embodiments, have at the sequence in the miRNAs in the binding site of miRNAs in the described specificity and the specificity and after described carrier being incorporated in the cell that comprises miRNAs in the described specificity, make miRNAs in the described specificity instruct enough complementarity at the described exogenous RNA molecule of described cracking site cracking.In other embodiments, cracking site can be positioned at the binding site at the specificity miRNAs, and cracking site can be between the sequence that suppresses sequence and the described exogenous object protein of encoding.In other embodiments, miRNAs is the Microrna of cell, viral Microrna or the two in the specificity.The Microrna of cell only can be expressed in neoplastic cell.The virus Microrna can be by the expressing viral that is selected from by the following group of forming: double-stranded DNA virus, single-stranded DNA viruses, diplornavirus, diplornavirus, strand (normal chain) virus, strand (minus strand) virus and retrovirus.
According to other embodiments, exogenous object protein is toxin.The group of the following composition of the optional freedom of described toxin: Ricin, ricin A chain, abrin, abrin A chain, diphtheria toxin A chain with and modified forms.In other embodiments, the group of the following composition of the optional freedom of described toxin: alpha toxin, saporin, Zea mays RIP, barley RIP, wheat RIP, corn RIP, rye RIP, flax RIP, shiga toxin, will are congratulated sample RIP, momordin, thymidine kinase, Pokeweed antiviral protein, are spent more white tree toxalbumin, Pseudomonas exotoxin, ETA, coli cytosine deaminase and modified forms thereof.
From specification sheets subsequently, objects and advantages of the present invention will be tangible.
The accompanying drawing summary
Exemplary and following figure is provided without limitation.
Fig. 1 is the synoptic diagram for the model of the biology generation of Microrna (miRNA) and activity.
Fig. 2 shows the synoptic diagram that the exogenous RNA molecule is activated by endogenous miRNA according to some embodiment, and wherein the inhibition sequence in the exogenous RNA molecule is positioned at the cracking site upstream of this exogenous RNA molecule.
Fig. 3 shows the synoptic diagram that the exogenous RNA molecule is activated by endogenous miRNA according to some embodiment, and wherein the inhibition sequence in the exogenous RNA molecule is positioned at the cracking site downstream of this exogenous RNA molecule.
Fig. 4 A is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises the AUG that is not positioned at same reading frame with the sequence of encoding exogenous target protein.
Fig. 4 B is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises not the Kozak consensus sequence (5 '-ACCAUGG-3 '-SEQ ID NO.25) that is positioned at same reading frame with the sequence of encoding exogenous target protein.
Fig. 4 C is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises 2 Kozak consensus sequences that are not positioned at same reading frame with the sequence of encoding exogenous target protein.
Fig. 5 A is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises AUG and the downstream terminator codon that is positioned at same reading frame.
Fig. 5 B is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises AUG and be used for downstream sorting signals or the proteolytic degradation signal of Subcellular Localization.
Fig. 5 C is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises AUG and coding can suppress the amino acid whose downstream sequence of biological function of downstream exogenous object protein.
Fig. 5 D is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, described inhibition sequence is positioned at the upstream of cracking site and comprises AUG, is positioned at downstream terminator codon and the downstream intron of same reading frame with AUG, wherein this exogenous RNA molecule is decay (nonsense-mediated decay, target NMD) of nonsense mediation.
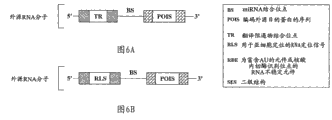
Fig. 6 A is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises binding site at translation repressor.
Fig. 6 B is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises RNA signal for locating for Subcellular Localization.
Fig. 6 C is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and is included as the element that is rich in AU or the unstable element (RNA destabilizing element) of the RNA of endonuclease recognition site.
Fig. 6 D is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the upstream of cracking site and comprises secondary structure.
Fig. 7 is the synoptic diagram that shows the example that the exogenous RNA molecule activated by endogenous miRNA according to some embodiment, wherein suppress sequence produce the secondary structure that hinders translation and wherein the cracking by miRNA produce IRES (internal ribosome entry site).
Fig. 8 A shows according to some embodiment to be increased in 5 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein other structure is IRES (internal ribosome entry site).
Fig. 8 B shows according to some embodiment to be increased in 5 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein other structure is stem-ring structure.
Fig. 8 C shows according to some embodiment to be increased in 5 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein other structure is tenuigenin polyadenylation element.
Fig. 8 D shows according to some embodiment to be increased in 5 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, wherein said other structure is to be bonded to each other and to impel described exogenous RNA molecule to form the nucleotide sequence of ring texture, particularly when the exogenous RNA molecule when cracking site is cleaved.
Fig. 9 A shows according to some embodiment to be increased in 5 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, wherein said other structure is by composition encoded polypeptides of the present invention, therefore wherein this polypeptide can be in conjunction with poly A and in conjunction with the sequence in the exogenous RNA and impel described exogenous RNA molecule to form ring texture, particularly when the exogenous RNA molecule when cracking site is cleaved.
Fig. 9 B shows according to some embodiment to be increased in 5 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, wherein said other structure is also can be in conjunction with the other RNA molecule of exogenous RNA molecule by composition coding of the present invention, and when the exogenous RNA molecule when cracking site is cleaved, provide cap by this other RNA molecule for it.
Fig. 9 C is the synoptic diagram of example that shows the other structure of the translation efficiency that reduces complete exogenous RNA molecule according to some embodiment, and wherein said other structure is to remove the cis acting type ribozyme of the cap sequence in the complete exogenous RNA molecule.
Figure 10 A shows the synoptic diagram of the sequence of very effective cis acting set hammer head ribozyme-snorbozyme (SEQ ID NO.63) [15].
Figure 10 B shows the synoptic diagram of the sequence of very effective cis acting set hammer head ribozyme-N117 (SEQ ID NO.64) [16].
Figure 11 A is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the cracking site downstream and comprises intron, and wherein said exogenous RNA molecule is the target of the decay (NMD) of nonsense mediation.
Figure 11 B is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the cracking site downstream and comprises binding site at translation repressor.
Figure 11 C is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the cracking site downstream and comprises RNA signal for locating for Subcellular Localization.
Figure 11 D is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the downstream of cracking site and is included as the element that is rich in AU or the unstable element of the RNA of endonuclease recognition site.
Figure 11 E is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the downstream of cracking site and comprises secondary structure.
Figure 12 A is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the sequence downstream of encoding exogenous target protein, and wherein said inhibition sequence produces the secondary structure that hinders translation.
Figure 12 B shows according to some embodiment to be increased in 3 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein said other structure is IRES (internal ribosome entry site).
Figure 12 C shows according to some embodiment to be increased in 3 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein said other structure is loop-stem structure.
Figure 12 D shows according to some embodiment to be increased in 3 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein said other structure is tenuigenin polyadenylation element.
Figure 13 A shows according to some embodiment to be increased in 3 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, and wherein said other structure is the nucleotide sequence that can be bonded to each other and impel described exogenous RNA molecule formation ring texture when the exogenous RNA molecule when cracking site is cleaved.
Figure 13 B shows according to some embodiment to be increased in 3 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, wherein said other structure is by the composition encoded polypeptides, wherein said polypeptide can be in conjunction with cap and in conjunction with the sequence in the described exogenous RNA and impel described exogenous RNA molecule to form ring texture, particularly when the exogenous RNA molecule when cracking site is cleaved.
Figure 13 C shows according to some embodiment to be increased in 3 ' synoptic diagram of the example of the other structure of the translation efficiency of the cleaved exogenous RNA molecule of end, wherein said other structure is by composition of the present invention coding and can be in conjunction with the exogenous RNA molecule and therefore for it provides the other RNA molecule of poly A, particularly when the exogenous RNA molecule when cracking site is cleaved.
Figure 13 D is the synoptic diagram of example that shows the other structure of the translation efficiency that reduces complete exogenous RNA molecule according to some embodiment, and wherein said other structure is to remove the cis acting type ribozyme of the poly A in the complete exogenous RNA molecule.
Figure 14 A is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, and described exogenous RNA molecule comprises two binding sites at different endogenous miRNA, wherein suppresses the upstream that sequence is positioned at described cracking site.
Figure 14 B is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, and described exogenous RNA molecule comprises two binding sites at identical endogenous miRNA, wherein suppresses the upstream that sequence is positioned at described cracking site.
Figure 14 C is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, and described exogenous RNA molecule comprises two binding sites at different endogenous miRNA, wherein suppresses the downstream that sequence is positioned at described cracking site.
Figure 14 D is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, and described exogenous RNA molecule comprises two binding sites at identical endogenous miRNA, wherein suppresses the downstream that sequence is positioned at described cracking site.
Figure 15 A is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, described exogenous RNA molecule has the inhibition sequence in the sequence downstream that is positioned at the encoding exogenous target protein, wherein said exogenous RNA molecule also comprises the initiator codon at the other binding site of miRNA and described other binding site upstream of the sequence upstream of encoding exogenous target protein, and wherein said initiator codon is not positioned at same reading frame with the sequence of encoding exogenous target protein.
Figure 15 B is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, described exogenous RNA molecule has the inhibition sequence in the sequence downstream that is positioned at the encoding exogenous target protein, and described exogenous RNA molecule also comprises the initiator codon at the other binding site of miRNA and described other binding site upstream of the sequence upstream of encoding exogenous target protein, and wherein said initiator codon is not positioned at same reading frame with the sequence of encoding exogenous target protein and wherein said exogenous RNA molecule also comprises the cis acting type ribozyme that is positioned at 5 ' end.
Figure 15 C is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, described exogenous RNA molecule comprises the sequence of encoding exogenous target protein between two miRNA binding sites, and comprise that also two are suppressed sequences, one at 5 ' end and another is at 3 ' end.
Figure 15 D is the synoptic diagram that shows the example of exogenous RNA molecule according to some embodiment, described exogenous RNA molecule comprises the sequence of encoding exogenous target protein between two different miRNA binding sites, and also comprise 2 and suppress sequences, one at 5 ' end and another is at 3 ' end.
Figure 16 A is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the cracking site downstream and can suppresses function for the RNA signal for locating of Subcellular Localization.
Figure 16 B is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the cracking site upstream and can suppresses function for the RNA signal for locating of Subcellular Localization.
Figure 16 C is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, and described inhibition sequence is positioned at the cracking site upstream and comprises AUG and coding can suppress amino acid whose downstream sequence for the function of the sorting signals of the Subcellular Localization of exogenous object protein.
Figure 16 D is the synoptic diagram that shows the example of the inhibition sequence in the exogenous RNA molecule according to some embodiment, described inhibition sequence is positioned at miRNA binding site downstream, and wherein said exogenous RNA molecule does not comprise the terminator codon in the downstream of starting codon of the sequence of encoding exogenous target protein.Suppress the aminoacid sequence of cracking that sequence encoding can suppress the peptide sequence of upstream coding, wherein this peptide sequence can be by the protease cracking in the target cell.
Figure 17 shows the synoptic diagram that composition of the present invention kills the purposes of the relevant cancer of the stomach cancer cells of the Burkitt lymphoma cancer cells that comprises endogenous miR-BART1, EBV-and nasopharyngeal carcinoma cancer cells according to some embodiment.
Figure 18 is the synoptic diagram that shows the example of the cell that the HIV-1 that uses composition of the present invention to kill to comprise endogenous hiv1-miR-N367 infects according to some embodiment.
Figure 19 shows according to some embodiment to use composition of the present invention to kill the synoptic diagram of the example of the metastatic breast cancer cell that comprises endogenous miR-10b.
Figure 20 shows according to some embodiment to use composition of the present invention to kill the synoptic diagram of the example of the cell that comprises endogenous miR-LAT.
Detailed Description Of The Invention
In following detailed description of the present invention, when use refers to term, for example: described, should, last is during with the former, it (for example refers to definite term mentioned above, when statement " this nucleotide sequence ", it refers to nucleotide sequence mentioned above rather than refers to nucleotide sequence mentioned above).In addition, in following detailed description of the present invention, each embodiment that is called other embodiments is defined as independent unit by it.
Be to spread all over that this specification sheets uses and should be according to the term of the following understanding of various embodiments below:
As referred to herein, term " polynucleotide molecule ", " oligonucleotide ", " polynucleotide ", " nucleic acid " and " Nucleotide " sequence are used in this article interchangeably.These terms refer to the form of independent fragment or as polymkeric substance or its heterozygote (hybrid) of deoxyribonucleotide (DNA), ribonucleotide (RNA) or its modified forms of the straight or branched of the component of bigger construct, strand, two strands, three chains.This term also comprises the RNA/DNA heterozygote.Polynucleotide can be for example DNA or RNA justice and antisense oligonucleotide or polynucleotide sequence arranged.DNA or RNA molecule can be such as but not limited to: complementary DNA (cDNA), genomic dna, synthetic DNA, recombinant DNA or its heterozygote or RNA molecule such as, for example mRNA, shRNA, siRNA, miRNA and similar molecule.Therefore, as used herein, term " polynucleotide molecule ", " oligonucleotide ", " polynucleotide ", " nucleic acid " and " Nucleotide " sequence be intended to refer to DNA and RNA molecule the two.These terms also comprise the oligonucleotide that is made of covalent linkage between naturally occurring base, sugar and nucleosides, and the oligonucleotide with part that the non-natural that similarly plays a role with separately naturally occurring part exists.
Term " polypeptide ", " peptide " and " protein " are used to refer to the polymkeric substance of amino-acid residue interchangeably by this paper.These term application are aminoacid polymerss of corresponding naturally occurring amino acid whose artificial chemical analog in wherein one or more amino-acid residues, and naturally occurring aminoacid polymers.
As referred to herein, term " complementarity " refers to the base pairing between the nucleic acid chains.As known in the art because the base pair of these interchains is by the non-covalent connection of two or three hydrogen bonds, so every chain of nucleic acid can with another chain complementation.Two Nucleotide that pass through the hydrogen bond connection on the relative complementary nucleic acid chain are called base pair.According to the base pairing of Wo Sen-Ke Like DNA, VITAMIN B4 (A) forms base pair with thymus pyrimidine (T) and guanine (G) with cytosine(Cyt) (C).In RNA, thymus pyrimidine is substituted by uridylic (U).Complementarity between two chains of nucleic acid can change according to the number (or per-cent) of the Nucleotide that forms base pair between these chains.For example, all Nucleotide and the complementary strand in every chain of " 100% complementarity " expression forms base pair.For example, Nucleotide and the complementary strand of 95% in every chain of " 95% complementarity " expression form base pair.The enough complementarity of term can comprise from about 30% to about 100% any complementary per-cent.
Term " construct " refers to artificial assembling or the isolated nucleic acid molecule that can be made of one or more nucleotide sequences as used herein, wherein these nucleotide sequences can be encoding sequence (sequence of coding end product just), regulating and controlling sequence, non-coding sequence or its any combination.The term construct for example comprises carrier but should not be considered as being confined to this.
" expression vector " refers to have with the heterology nucleic acid fragment (carrier that for example, DNA) is incorporated in the foreign cell and expresses the ability of described heterology nucleic acid fragment in described foreign cell.In other words, expression vector comprises the nucleotide sequence/fragment (for example, DNA, mRNA, tRNA, rRNA) that can be transcribed.Many viruses, protokaryon and carrier for expression of eukaryon are known and/or commercially available.The selection of suitable expression vector is in those skilled in the art's the ken.
Term " upstream " and " downstream " refer at nucleotide sequence such as, the relative position in dna sequence dna or the RNA sequence for example as used herein.As everyone knows, nucleotide sequence has the 5 ' end and 3 of so-called carbon at the sugar on the Nucleotide skeleton (ribodesose or ribose) ring ' hold.Therefore, with respect to the position that lists at nucleotides sequence, the term downstream refers to the zone towards 3 of sequence ' end.The term upstream refers to the zone towards 5 of chain ' end.
Term " promoter element ", " promotor " or " promoter sequence " refer to be usually located at 5 of encoding sequence ' end (just, before it, be located thereon trip) and serve as switch, the nucleotide sequence that the activated code sequence is expressed as used herein.If encoding sequence is activated, then it should be expressed as and be transcribed.Transcribe and generally include from the synthetic RNA molecule of encoding sequence (such as, mRNA for example).Therefore, promotor is used as transcriptional regulatory element and also provides and is used for starting the site that encoding sequence is transcribed into mRNA.Promotor can derive from natural origin fully, or the different elements that origin comes from the different promoters that occurring in nature finds constitutes, or even comprises synthetic Nucleotide section.It will be understood by those skilled in the art that different promotors can instruct gene in different tissues or cell type, or in the different etap, or in response to different envrionment conditionss or with different expression level expression.Most of the time, cause gene expression promoter in most cell types to be commonly called " composition promotor ".Drive gene expression promoter in particular organization and be called as " tissue-specific promoter ".
As referred to herein, the recombinant RNA molecule that term " exogenous RNA molecule " refers to be introduced in the target cell and/or expresses in target cell.The exogenous RNA molecule can be complete (full-length molecule just) or can be cleaved at one or more cracking sites in cell.
As used herein, term " target protein " and " exogenous object protein " are used interchangeably.The peptide sequence of being translated by the exogenous RNA molecule in these term phalangeal cells.In certain embodiments, described peptide sequence can be one or more different protein or fusion roteins.
As used herein, term " miRNAs in the specificity " and " specificity miRNA " use interchangeably.Microrna (miRNA) molecule/sequence in these term phalangeal cells.MiRNAs can be by the genome encoding (cell miRNA) of cell in the specificity, and/or by foreign gene group resident in the cell, such as for example, by encoding viral resident in the cell (viral miRNA).Specificity miRNA before the exogenous RNA molecule is introduced into target cell/be present in the target cell before being expressed.
Term " expression " refers to produce the end product molecule of expectation in target cell as used herein.The end product molecule can be RNA molecule for example; Peptide or protein; And similar molecule; Or its combination.
As referred to herein, term " opening code-reading frame " (" ORF ") refers to comprise the coding region of starting codon with terminator codon.
As referred to herein, term " Kozak sequence " is well known in the art and refers on the mRNA molecule by the sequence of rrna as translation initiation site identification.Term " Kozak consensus sequence ", " Kozak consensus " or " Kozak sequence " are the sequences that is present on the eukaryotic mrna and has consensus (gcc) gccRccAUGG (SEQ ID NO.24), wherein, R is purine (VITAMIN B4 or guanine), be positioned at three the base places, (AUG) upstream of starting codon, start codon (AUG) is another ' G ' afterwards.In certain embodiments, the Kozak sequence has sequence RNNAUGG (SEQ ID NO.83).
As used herein, term " introducing " and " transfection " are used interchangeably and are referred to molecule such as for example nucleic acid, polynucleotide molecule, carrier and similar molecular transfer in target cell, and more particularly transfer to the inside in the space that the film of target cell seals.These molecules can be by any method known to those skilled in the art by " introducing " in target cell, for example, by people such as Sambrook, Molecular Cloning:A Laboratory Manual, Cold Spring Harbor Laboratory Press, the method of New York (2001) instruction, the content of the document is incorporated into this paper by reference.Molecule " introducing " is comprised such as, but not limited to: heat shock, calcium phosphate transfection, PEI transfection, electroporation, lipofection, transfection reagent, virus-mediated transfer and similar approach or its combination to intracellular method.The transfection of cell can for example be carried out such as people's cell, zooblast, vegetable cell and similar cell at the cell in any source of any kind.These cells can be the interior cell that exists of cell, clone, organism and the similar cells of isolated cells, tissue culture.
" kill " about the term of cell/cell mass and to refer to comprise and to cause the operation of any kind of the death of cell/cell mass.
As referred to herein, term " treatment disease " or " treatment illness " refer to use a kind of composition, described composition comprises and alleviates the symptom relevant with disease effectively with the severity that reduces disease or cure described disease or at least a reagent (it for example can be, one or more polynucleotide molecules, one or more carriers, one or more material/compositions and similar reagents) to prevent described disease to take place.Use and to comprise any route of administration.
Term " detection ", " diagnosis " refer to detect the detection of disease, symptom, illness, pathological conditions or normal condition (pathological or normal condition); To disease, symptom, illness, pathological conditions classification; Determine the seriousness of disease, symptom, illness, pathological conditions; The progress of monitoring of diseases, symptom, illness, pathological conditions; Predict the method for the prospect of its result and/or recovery.
1.
The basic structure of composition of the present invention
According to some embodiment, provide to be used for the composition that only cell of miRNAs is expressed exogenous object protein in comprising specificity.Endogenous miRNA can be the miRNA of any kind of existence in the miRNA of cell, viral miRNA and/or the cell.Exogenous object protein can be peptide or the protein of any kind, for example such as toxin.
According to some embodiment, composition of the present invention can comprise one or more polynucleotide molecules, for example such as dna molecular, RNA molecule or the two.
In certain embodiments, described composition comprises the exogenous RNA molecule or this molecule of encoding, and described exogenous RNA molecule is the RNA molecule that comprises following at least sequence:
A) sequence of encoding exogenous target protein;
B) can suppress the inhibition sequence that this exogenous object protein is expressed; With
C) be designed to specificity in the ripe miRNA chain of miRNAs have and make that miRNAs instructs described exogenous RNA molecule at the binding site of enough complementarity of cracking site cracking in the described specificity.This cracking site is designed between the sequence that suppresses sequence and this exogenous object protein of encoding.
Therefore, only in cell, exist in the specificity under the miRNAs, the exogenous RNA molecule this cracking site by described specificity in miRNAs cracking and suppress sequence and separate with the sequence of encoding exogenous target protein, and described exogenous object protein can be expressed.For example, this is illustrated in Fig. 2 and 3.
According to some embodiment, select in the specificity miRNAs may with its relevant for the expression in the particular cell types of target cell and/or determine according to described expression.Therefore, be chosen in the mechanism that the interior miRNAs of the specificity of expressing in the particular cell types can be provided for the described exogenous object protein of (target cell) targeted expression in the cell type of selected selection thus.Described specific cell can be selected from such as, but not limited to the cells that infected by virus or other morbid substances; The cell of optimum or malignant cell, the immune component of expression.Specificity can be modified to by the binding site with the exogenous RNA molecule of described composition with specificity in the ripe miRNA chain of miRNAs have and make that miRNAs instructs external source mRNA molecule enough complementarity of cracking in target cell to realize in the described specificity.
Known in the art is that the mRNA that does not contain cap or poly A tract bar still can translated protein.In mammalian cell, add cap and make the mRNA translation be increased to 35-50 doubly to add poly (A) tail and make the mRNA translation be increased to 114-155 doubly [6].Poly in the mammalian cell (A) tail only makes the transformation period of functional mRNA be increased to 2.6 times and cap only makes the functional mRNA transformation period be increased to 1.7 times [6].
It is also known that in this area, some protein can with in addition the concentration of a protein of each cell cell is brought into play biological action.For example, but reported the single Ricin of the cytosol that arrives cell or the albumen cell killing [3,4] of abrin.In addition, be incorporated in the cell single diphtheria toxin Segment A (DTA) but albumen cell killing [5].In certain embodiments, exogenous object protein can be any protein or peptide, such as such as but not limited to Ricin, abrin, diphtheria toxin and similar toxin or its combination.
According to some embodiment, exogenous object protein can be the polypeptide for the syzygy of two kinds of protein, and described polypeptide can have cracking site between two kinds of protein, allows two kinds of protein to separate in cell.For example, exogenous object protein can be the fusion rotein of Ricin and DTA, and wherein this fusion rotein can cause forming DTA separately and the protein of Ricin by the cracking of for example specific protease in cell.In certain embodiments, described exogenous object protein can be two kinds of different protein can being expressed by described composition.For example, two kinds of different exogenous object protein of external source purpose RNA codified, such as, for example Ricin and DTA.
2.
Structure with exogenous RNA molecule of the inhibition sequence that is positioned at the cracking site upstream
2.1.
Be positioned at the structure of the inhibition sequence of cracking site upstream
According to some embodiment, the inhibition sequence in the exogenous RNA molecule can be positioned at downstream or the upstream of cracking site.The structure of the inhibition sequence that is arranged in exogenous RNA molecule cracking site upstream has been described in this part.This for example is illustrated among Fig. 2.
According to some embodiment, the inhibition sequence that is positioned at the cracking site upstream for example can be, initiator codon.The sequence of described initiator codon and the described exogenous object protein of coding is not in the same reading frame, makes described initiator codon can cause the phase shift mutation at described exogenous object protein, and the encoding sequence of described exogenous object protein is positioned at the downstream.This for example is illustrated in, among Fig. 4 A.In one embodiment, initiator codon can be positioned at the Kozak consensus sequence.In addition, also can use maintenance to serve as the adorned Kozak consensus sequence of the ability of translation initiation thing (initiator).For example, referring to Fig. 4 B.In certain embodiments, people's Kozak consensus sequence be 5 '-ACCAUGG-3 ' (SEQ ID NO.25) and initiator codon be 5 '-AUG-3 '.
In certain embodiments, initiator codon can be positioned at the TISU motif or have one or more TISU motifs.TISU (short 5 ' UTR translation initiation thing) motif is different from Kozak consensus [38] because the guidance of its uniqueness has the ability that the mRNA of 5 very short ' UTR effectively and exactly begins to translate.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream can have a plurality of initiator codons, and wherein the sequence of each initiator codon and encoding exogenous target protein is not in same reading frame.These initiator codons can cause the phase shift mutation of exogenous object protein, and the encoding sequence of described exogenous object protein is positioned at the downstream.In addition, each initiator codon can be positioned at the Kozak consensus sequence or keep serving as the Kozak consensus sequence of modification of the ability of translation initiation thing.For example, referring to Fig. 4 C.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream can comprise initiator codon.The exogenous RNA molecule also can comprise terminator codon between the starting codon of the sequence of initiator codon and encoding exogenous target protein, wherein said terminator codon and initiator codon are in the same reading frame.In this embodiment, produced the upstream opening code-reading frame (uORF) of the translation efficiency of the downstream sequence that can reduce the described exogenous object protein of coding.For example, referring to Fig. 5 A.In certain embodiments, terminator codon can be, for example 5 '-UAA-3 ' or 5 '-UAG-3 ' or 5 '-UGA-3 '.
In certain embodiments, strong stem and ring structure can be positioned at upstream or the downstream of ORF, in the upstream or the location downstream that are the target sequence (cracking site) of miRNA.The generation of these stems and ring can be helpful under following situation: although wherein arrived terminator codon, ribosomal small subunit with on the mRNA does not separate, and continues scanning mRNA.Ribosomal small subunit can not be opened firm RNA secondary structure.In addition, when these stems were positioned at the downstream of target sequence with ring, it is the degraded of for example being undertaken by the XRN1 exoribonuclease to cleaved mRNA capable of blocking also.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream can comprise the nucleotide sequence that initiator codon and coding are used for the sorting signals of Subcellular Localization.Described nucleotide sequence can be arranged in the downstream of initiator codon and described nucleotide sequence and initiator codon and be positioned at same reading frame.In certain embodiments, the Subcellular Localization of the exogenous object protein that is instructed by sorting signals can suppress the biological function of target protein.The sorting signals that is used for Subcellular Localization can be such as but not limited to: plastosome sorting signals, nuclear sorting signals, endosome sorting signals, lysosome sorting signals, peroxysome sorting signals, ER sorting signals and similar sorting signals.The sorting signals that is used for Subcellular Localization can be (people's alkylphosphonic acid carboxylic acid otan (alkyl dihydroxyacetonephosphate) synthase) peroxysome target signal 2[(R/K for example) (L/V/I) X
5(Q/H) (L/A)] (SEQ ID NO.26) or H
2N----RLRVLSGHL (SEQ ID NO.27) [28].This for example is illustrated in, among Fig. 5 B.
In another embodiment of the invention, the inhibition sequence that is positioned at the cracking site upstream can comprise the nucleotide sequence of initiator codon and proteins encoded degraded signal.Described nucleotide sequence is positioned at the downstream of initiator codon, and wherein said nucleotide sequence and initiator codon are positioned at same reading frame.The proteolytic degradation signal can be such as, but not limited to ubiquitin degraded signal.For example, referring to Fig. 5 B.
In another embodiment of the invention, the nucleotide sequence that the inhibition sequence that is positioned at the cracking site upstream can be designed to comprise initiator codon and be positioned at described initiator codon downstream, described nucleotide sequence and described initiator codon and be positioned at same reading frame with the sequence of encoding exogenous target protein, wherein when described nucleotide sequence coded aminoacid sequence was fused to exogenous object protein, the biological function of described exogenous object protein was suppressed.For example, referring to Fig. 5 C.
In another embodiment of the invention, the inhibition sequence that is positioned at the cracking site upstream can comprise initiator codon, and the exogenous RNA molecule also can comprise the terminator codon in described initiator codon downstream, and wherein said terminator codon and described initiator codon are in same reading frame.In addition, described exogenous RNA molecule also can comprise the intron in described terminator codon downstream, and wherein said exogenous RNA molecule is the target [29] of decay (NMD) of the nonsense mediation of the described exogenous RNA molecule of degradable.For example, referring to Fig. 5 D.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream can comprise can the combining translation aporepressor sequence.In certain embodiments, translation repression albumen is endogenous translation repression albumen.In certain embodiments, translation repression albumen can be by described composition coding.Described translation repression albumen can reduce the translation efficiency [24] of exogenous object protein directly or indirectly.For example, sequence that can the combining translation aporepressor includes but not limited to the sequence (5 '-UGGAGCAGAGGCUCUGGCAGCUUUUGCAGCG-3 ') (SEQ ID NO.28) [25] in conjunction with the SMAUG aporepressor.For example, referring to Fig. 6 A.
In another embodiment, being positioned at the RNA signal for locating that the inhibition sequence of cracking site upstream can comprise for Subcellular Localization (comprises, for example, altogether translation input (co-translational import)) or endogenous miRNA binding site, wherein the Subcellular Localization of exogenous RNA molecule can suppress the translation of exogenous object protein and can shorten transformation period of exogenous RNA molecule.The RNA signal for locating can be such as but not limited to, and is used for following RNA signal for locating: myelinization periphery (myelinating periphery), myelin compartment (myelin compartment), plastosome, thin plate leading edge (leading edge of the lamella), nuclear cortical cytoplasm [22] or similar position.For example, the RNA signal for locating can be for the myelinization periphery RNA signal for locating 5 '-GCCAAGGAGCCAGAGAGCAUG-3 ' (SEQ ID NO.29) or 5 '-GCCAAGGAGCC-3 ' (SEQ ID NO.30) [27].For example, referring to Fig. 6 B.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream can comprise the unstable element of the RNA that can stimulate the exogenous RNA molecular degradation.The unstable element of RNA for example can be, is rich in element (ARE), the endonuclease recognition site or similar of AU.The element that is rich in AU can be that for example length is at least about the element that is rich in AU of 35 Nucleotide.For example, the element that is rich in AU can be 5 '-AUUUA-3 ' (SEQ ID NO.31), 5 '-UUAUUUA (U/A) (U/A)-3 ' (SEQ ID NO.32) or 5 '-AUUU-3 ' (SEQ ID NO.33) [26].For example, referring to Fig. 6 C.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream can comprise the secondary structure that can form the translation efficiency that can reduce the downstream exogenous object protein.In certain embodiments, the folding free energy of secondary structure can be lower than-30kcal/mol (for example ,-50kcal/mol ,-80kcal/mol) and therefore this secondary structure is enough to hinder the starting codon of downstream area that the scanning rrna arrives the encoding exogenous target protein.For example, referring to Fig. 6 D.
In other embodiments, the inhibition sequence that is positioned at the cracking site upstream can comprise the nucleotide sequence of next-door neighbour's cracking site located upstream, wherein said nucleotide sequence can be in conjunction with the nucleotide sequence that is close to the cracking site downstream location to form secondary structure, and wherein said secondary structure can reduce the translation efficiency of downstream exogenous object protein directly or indirectly.
The folding free energy of secondary structure can be lower than-30kcal/mol (for example ,-50kcal/mol ,-80kcal/mol) and therefore this secondary structure is enough to hinder the scanning rrna and arrives starting codon of exogenous object protein.In another embodiment, described cracking site can be arranged in the ring zone of strand zone or secondary structure, and wherein said strand zone or described ring zone can be such as but not limited to the zone that length is at least about 15 Nucleotide.In another embodiment, the exogenous RNA molecule also can comprise the sequence of internal ribosome entry site (IRES) of the sequence upstream of cracking site downstream and encoding exogenous target protein, wherein said IRES sequence at the exogenous RNA intramolecularly of cracking than with better function at complete exogenous RNA intramolecularly.In another embodiment, at least a portion of IRES sequence can be positioned at the nucleotide sequence of the downstream location of next-door neighbour's cracking site.For example, referring to Fig. 7.
The IRES sequence can be selected from, such as but not limited to the IRES of pico+ribonucleic acid+virus, the IRES of foot and mouth disease (foot-and-mouth disease) virus, the IRES of encephalomyocarditis virus, the IRES of hepatitis A virus, the IRES of hepatitis C virus, human rhinovirus's IRES, the IRES of poliovirus, the IRES of swine vesicular disease virus, the IRES of the Brassica 2 et 4 of marmor upsilon group (turnip mosaic potyvirus), the IRES of human fibroblastic growth factor 2mRNA, the IRES of pestivirus, the IRES of leishmania RNA viruses, the IRES of Moloney murine leukemia virus, the IRES of ERC group virus 14, the IRES of foot and mouth disease virus (aphthovirus), the human immunoglobulin heavy chain is in conjunction with the IRES of protein mRNA, the IRES (Drosophila Antennapedia mRNA IRES) of the mRNA of the gene of fruit bat control feeler, the IRES of human fibroblastic growth factor 2mRNA, the IRES of hepatitis G virus, the IRES of tobacco mosaic virus (TMV), the IRES of vascular endothelial growth factor mRNA, the IRES of B group Coxsackie virus, the IRES of c-myc proto-oncogene mRNA, the IRES of people MYT2mRNA, the IRES of human parechovirus's 1 C-type virus C, the IRES of human parechovirus's 2 C-type virus Cs, the IRES of eukaryotic initiation factor 4GI mRNA, little amber stinkbug (Plautia stali) enterovirus IRES, mouse Tai Leshi encephalomyelitis virus IRES, bovine enteroviruses IRES, the IRES that connects protein 43 mRNA, the IRES of homeodomain protein Gtx mRNA, the IRES of AML1 transcription factor mRNA, the IRES of NF-κ B supressor mRNA, the IRES of the chain survivin mRNA of X, the IRES of cricket paralysis virus RNA, the IRES of p58 (PITSLRE) protein kinase mRNA, the IRES of ornithine decarboxylase mRNA, the IRES that connects protein 32 mRNA, bovine viral diarrhea virus IRES, the IRES of insulin-like growth factor I receptor mRNA, the IRES of human 1 type immunodeficiency virus gag gene, classic Pestivirus suis IRES, the IRES of kaposi sarcoma-associate herpesvirus, be selected from the short IRES in random oligonucleotide library, the viral IRES of Jembrana disease, the IRES of apoptosis protease-activating factor 1mRNA, the IRES of rhopalosiphum padi (Rhopalosiphum padi) virus, the IRES of cationic amino acid transporter mRNA, the IRES of the leading factor 2mRNA of human insulin-like growth factor II (human insulin-like growth factor II leader 2 mRNA IRES), the IRES of flagellate virus, the IRES of Smad5 mRNA, the IRES of the prompt Shen virus-1talfan of pig, fruit bat does not have the IRES of mao mRNA, the IRES of hSNM1 mRNA, the IRES of Cbfa1/Runx2mRNA, the IRES of Epstein epstein-Barr virus, the IRES of the withered and yellow ring spot virus of lotus, the IRES of rat pituitary Hou Yejiayasu V1b receptor mrna or the IRES of people hsp70mRNA.
2.2.
Can be increased in the other structure of the translation efficiency of the cleaved exogenous RNA molecule of 5 ' end
This part has been described the other embodiments of structure of the translation efficiency of the exogenous RNA molecule that can increase cracking in detail, and the exogenous RNA molecule of wherein said cracking is cleaved at the cracking site at 5 ' end place.
According to some embodiment, the exogenous RNA molecule can comprise the sequence of internal ribosome entry site (IRES) sequence of the uniqueness of the sequence upstream of containing next-door neighbour's encoding exogenous target protein, and the IRES sequence of wherein said uniqueness has increased the translation efficiency of exogenous object protein in the exogenous RNA molecule of cracking.For example, referring to Fig. 8 A.
In another embodiment, the exogenous RNA molecule can comprise the nucleotide sequence of uniqueness in the sequence downstream of next-door neighbour's encoding exogenous target protein, and stem-ring structure that the nucleotide sequence of wherein said uniqueness comprises unique stem-ring structure and wherein said uniqueness can increase the translation efficiency of exogenous object protein in the exogenous RNA molecule of cracking and the transformation period of exogenous RNA molecule directly or indirectly.Stem-the ring structure of described uniqueness can be, such as but not limited to human histone gene 3 '-conservative loop-stem structure or its functional derivatives of UTR.Human histone gene 3 '-the conservative loop-stem structure of UTR for example can be, be 5 '-GGCUCUUUUCAGAGCC-3 ' (SEQ ID NO.34).For example, referring to Fig. 8 B.
In other embodiments, the exogenous RNA molecule can comprise the nucleotide sequence of uniqueness in downstream of the sequence of next-door neighbour's encoding exogenous target protein, and the nucleotide sequence of wherein said uniqueness comprises the tenuigenin polyadenylation element of the transformation period of the translation efficiency of exogenous object protein in the exogenous RNA molecule that can increase cracking directly or indirectly and exogenous RNA molecule.Tenuigenin polyadenylation element can be such as, but not limited to: 5 '-UUUUAU-3 ' (SEQ ID NO.35), 5 '-UUUUUAU-3 ' (SEQ ID NO.36), 5 '-UUUUAAU-3 ' (SEQ ID NO.37), 5 '-UUUUUUAUU-3 ' (SEQ ID NO.38), 5 '-UUUUAUU-3 ' (SEQ ID NO.39) or 5 '-UUUUUAUAAAG-3 ' (SEQ ID NO.40) [23].
In certain embodiments, composition of the present invention also comprises the polynucleotide sequence of coding people's tenuigenin polyadenylation element conjugated protein (hCPEB) or it is used for expressing at any cell the homologue of hCPEB.For example, referring to Fig. 8 C.
In other embodiments, the exogenous RNA molecule can comprise the nucleotide sequence of the uniqueness of the sequence upstream that is positioned at cracking site downstream and encoding exogenous target protein, and the nucleotide sequence of wherein said uniqueness can be in conjunction with the sequence in the sequence downstream that is positioned at the encoding exogenous target protein.In this embodiment, the exogenous RNA molecule of cracking can produce the ring structure of the translation efficiency of exogenous object protein in the described exogenous RNA molecule that can increase cracking.For example, referring to Fig. 8 D.
In another embodiment, the exogenous RNA molecule can comprise the nucleotide sequence of the uniqueness of the sequence upstream that is positioned at cracking site downstream and encoding exogenous target protein.The nucleotide sequence of described uniqueness can bound energy enough directly or indirectly in conjunction with unique polypeptide of poly (A) tail in the exogenous RNA molecule of cracking.Described unique polypeptide also can be by composition coding of the present invention.In this embodiment, the exogenous RNA molecule of described unique polypeptide and cracking can produce the ring structure that can increase the translation efficiency of exogenous object protein described in the exogenous RNA of the cracking molecule.For example, referring to Fig. 9 A.
In another embodiment, composition of the present invention also can comprise other polynucleotide sequence, this other polynucleotide sequence coding 5 ' end comprises can be in conjunction with the other RNA molecule of the nucleotide sequence of the uniqueness of the sequence upstream that is positioned at cracking site downstream and encoding exogenous target protein.The expression of the polynucleotide sequence that this is other can be by for example based on the promoters driven of polymerase II.In certain embodiments, composition of the present invention also can comprise can influence this other RNA molecule directly or indirectly in the cracking component of the cracking of the nucleotide sequence location downstream of described uniqueness.Described cracking component for example can be:
(a) be positioned at the nucleotide sequence of the described other intramolecular uniqueness of RNA, the nucleotide sequence of wherein said uniqueness can be but be not limited to: endonuclease recognition site, endogenous miRNA binding site, cis acting type ribozyme, palindrome termination element or miRNA sequence; Or
(b) by the inhibitory RNA of the uniqueness of composition of the present invention coding, the inhibitory RNA of wherein said uniqueness can be but be not limited to: the RNA of Microrna (miRNA), lasso trick form, short hairpin RNA (shRNA), siRNA expression structure territory, sense-rna, double-stranded RNA (dsRNA), siRNA (siRNA) or ribozyme.
In this embodiment, described other RNA molecular energy can provide the cap sequence that can increase the translation efficiency of exogenous object protein described in the exogenous RNA of the cracking molecule in conjunction with the exogenous RNA molecule of cracking and for it.For example, referring to Fig. 9 B.
In certain embodiments, can introduce the vpg recognition sequence, wherein after cracking, 5 ' cracking end comprises the vpg recognition site.Vpg albumen can be in conjunction with the vpg recognition sequence, thereby replaces cap.Vpg albumen can be by composition of the present invention or by the ORF coding that suppresses sequence.
In certain embodiments, and do not expect to be fettered ground by theoretical or mechanism, it is favourable using cis acting type ribozyme, but because comprises its other RNA molecule self cracking [15].Cis acting type ribozyme can be such as, but not limited to very effective cis acting set hammer head ribozyme: snorbozyme[15] or N117[16].Referring to Figure 10 A, 10B.
In another embodiment, the exogenous RNA molecule also can comprise the nucleotide sequence of the sequence upstream of next-door neighbour's encoding exogenous target protein, and wherein said nucleotide sequence comprises the loop-stem structure of the degraded that can increase the exogenous RNA of cracking molecule.In one embodiment, described loop-stem structure be human histone gene 3 '-the conservative loop-stem structure of UTR (5 '-GGCUCUUUUCAGAGCC-3 '-SEQ ID NO.34) or its functional derivatives.
2.3.
Can reduce the other structure of the translation efficiency of complete exogenous RNA molecule
The multiple embodiments of other structure has been described in this part, and wherein these other structures can reduce the translation efficiency (just, before described exogenous RNA molecule is cleaved) of complete exogenous RNA molecule.
In certain embodiments, described composition also can comprise can realize the exogenous RNA molecule directly or indirectly in the specific cleavage component of the cracking that is positioned at the position that suppresses the sequence upstream, and wherein said inhibition sequence is positioned at the cracking site upstream.Described specific cracking component for example can be:
(a) be positioned at the intramolecular specific nucleic acid sequence of exogenous RNA, wherein said specific nucleic acid sequence can be such as but not limited to: endonuclease recognition site, endogenous miRNA binding site, cis acting type ribozyme or miRNA sequence; Perhaps
(b) by the specific inhibitory RNA of composition coding of the present invention, wherein said specific inhibitory RNA can be such as but not limited to: the RNA of Microrna (miRNA), lasso trick form, short hairpin RNA (shRNA), siRNA expression structure territory, sense-rna, double-stranded RNA (dsRNA), siRNA (siRNA) or ribozyme.
In this embodiment, described specific cleavage component can be removed the cap sequence in the complete exogenous RNA molecule, to reduce the translation efficiency of exogenous object protein in the complete exogenous RNA molecule.For example, referring to Fig. 9 C.
In another embodiment, the inhibition sequence that is positioned at the cracking site upstream also can comprise one or more initiator codons, wherein the sequence of each initiator codon and encoding exogenous target protein not in same reading frame and wherein each in these initiator codons be positioned at the Kozak consensus sequence.
3.
Structure with exogenous RNA molecule of the inhibition sequence that is positioned at the cracking site downstream
3.1.
Be positioned at the structure of the inhibition sequence in cracking site downstream
According to some embodiment, the inhibition sequence in the exogenous RNA molecule can be positioned at downstream or the upstream of cracking site.This part has been described and has wherein been suppressed the embodiment that sequence is arranged in the cracking site downstream of exogenous RNA molecule.For example, referring to Fig. 3.
In certain embodiments, the inhibition sequence that is positioned at the cracking site downstream can comprise for example intron.Therefore the exogenous RNA molecule can become the target [29] of decay (NMD) of the nonsense mediation of degraded exogenous RNA molecule.For example, referring to Figure 11 A.
In one embodiment, the inhibition sequence that is positioned at the cracking site downstream can comprise can the combining translation aporepressor sequence, wherein said translation repression albumen is endogenous translation repression albumen or by described composition coding, and wherein said translation repression albumen can reduce the translation efficiency [24] of exogenous RNA intramolecularly exogenous object protein directly or indirectly.Sequence that can the combining translation aporepressor can be the sequence (5 '-UGGAGCAGAGGCUCUGGCAGCUUUUGCAGCG-3 ') (SEQ ID NO.28) [25] that for example includes but not limited in conjunction with the smaug aporepressor.For example, referring to Figure 11 B.
In another embodiment, being positioned at the RNA signal for locating that the inhibition sequence in cracking site downstream can comprise for Subcellular Localization (comprises, translation is imported altogether) or endogenous miRNA binding site, the Subcellular Localization of wherein said exogenous RNA molecule can suppress the translation of exogenous object protein and can reduce the transformation period of exogenous RNA molecule.The RNA signal for locating can comprise such as but not limited to being used for following RNA signal for locating: myelinization periphery, myelin compartment, plastosome, thin plate leading edge or nuclear cortical cytoplasm [22].The RNA signal for locating can be such as but not limited to for the myelinization periphery RNA signal for locating 5 '-GCCAAGGAGCCAGAGAGCAUG-3 ' (SEQ ID NO.29) or 5 '-GCCAAGGAGCC-3 ' (SEQ ID NO.30) [27].For example, referring to Figure 11 C.
In another embodiment, the inhibition sequence that is positioned at the cracking site downstream can comprise the unstable element of the RNA that can stimulate the exogenous RNA molecular degradation, and wherein the unstable element of RNA is element (ARE) or the endonuclease recognition site that is rich in AU.The element that is rich in AU can be the element that is rich in AU that is at least about 35 Nucleotide such as, but not limited to length.The element that is rich in AU for example can be, 5 '-AUUUA-3 ' (SEQ ID NO.31), 5 '-UUAUUUA (U/A) (U/A)-3 ' (SEQ ID NO.32) or 5 '-AUUU-3 ' (SEQ ID NO.33) [26].For example, referring to Figure 11 D.
In another embodiment, the inhibition sequence that is positioned at the cracking site downstream can comprise the sequence of the secondary structure that can form the translation efficiency that can reduce the upstream exogenous object protein.For example, referring to Figure 11 E.
What in another embodiment, the inhibition sequence that is positioned at the cracking site downstream can comprise next-door neighbour's cracking site downstream can be in conjunction with the nucleotide sequence of next-door neighbour's cracking site located upstream to form the sequence of secondary structure.Described secondary structure can reduce the translation efficiency of upstream foreign protein directly or indirectly.In certain embodiments, the folding free energy of secondary structure can be lower than-30kcal/mol (for example ,-50kcal/mol ,-80kcal/mol) and therefore this secondary structure is enough to hinder the terminator codon that the scanning rrna arrives exogenous object protein.In another embodiment, described cracking site is arranged in the ring zone of strand zone or secondary structure, and wherein said strand zone or described ring zone can be the zones that is at least about 15 Nucleotide such as, but not limited to length.For example, referring to Figure 12 A.
3.2.
The translation that can be increased in the cleaved exogenous RNA molecule in the cracking site place at 3 ' end place is imitated The other structure of rate
The embodiment of other structure has been described in this part, and wherein these other structures can increase the translation efficiency of the exogenous RNA molecule of cracking, and the exogenous RNA molecule of wherein said cracking is cleaved at the cracking site place at 3 ' end place.
In certain embodiments, the exogenous RNA molecule can comprise the sequence of internal ribosome entry site (IRES) sequence of the uniqueness of the sequence upstream with next-door neighbour's encoding exogenous target protein, and the IRES sequence of wherein said uniqueness can increase the translation efficiency of exogenous object protein in the exogenous RNA molecule of cracking.For example, referring to Figure 12 B.
In another embodiment of the invention, the exogenous RNA molecule can comprise the nucleotide sequence of uniqueness in the sequence downstream of next-door neighbour's encoding exogenous target protein, and stem-ring structure that the nucleotide sequence of wherein said uniqueness comprises unique loop-stem structure and wherein said uniqueness can increase the translation efficiency of exogenous object protein of exogenous RNA molecule of cracking and the transformation period of exogenous RNA molecule directly or indirectly.The loop-stem structure of described uniqueness can be such as, but not limited to human histone gene 3 '-conservative loop-stem structure or its functional derivatives of UTR.Human histone gene 3 '-the conservative loop-stem structure of UTR is 5 '-GGCUCUUUUCAGAGCC-3 ' (SEQ ID NO.34).For example, referring to Figure 12 C.
In one embodiment of the invention, the exogenous RNA molecule of describing in 3.1 or 1 part can comprise the nucleotide sequence of uniqueness in downstream of the sequence of next-door neighbour's encoding exogenous target protein, and the nucleotide sequence of wherein said uniqueness comprises the tenuigenin polyadenylation element of the transformation period of the translation efficiency of exogenous object protein in the exogenous RNA molecule that can increase cracking directly or indirectly and exogenous RNA molecule.Tenuigenin polyadenylation element can be such as, but not limited to 5 '-UUUUAU-3 ' (SEQ ID NO.35), 5 '-UUUUUAU-3 ' (SEQ ID NO.36), 5 '-UUUUAAU-3 ' (SEQ ID NO.37), 5 '-UUUUUUAUU-3 ' (SEQ ID NO.38), 5 '-UUUUAUU-3 ' (SEQ ID NO.39) or 5 '-UUUUUAUAAAG-3 ' (SEQ ID NO.40) [23].Composition of the present invention also can comprise the polynucleotide sequence of coding people's tenuigenin polyadenylation element conjugated protein (hCPEB) or it is for the homologue of expressing hCPEB at any cell.For example, referring to Figure 12 D.
In certain embodiments, the exogenous RNA molecule can comprise the nucleotide sequence of the uniqueness in the sequence downstream that is positioned at cracking site upstream and encoding exogenous target protein, and the nucleotide sequence of wherein said uniqueness can be in conjunction with the sequence of the sequence upstream that is positioned at the encoding exogenous target protein.In this embodiment, the exogenous RNA molecule of cracking can produce the ring structure that can increase the translation efficiency of exogenous object protein in the exogenous RNA of the cracking molecule.For example, referring to Figure 13 A.
In another embodiment, the exogenous RNA molecule can comprise the nucleotide sequence of the uniqueness in the sequence downstream that is positioned at cracking site upstream and encoding exogenous target protein.The nucleotide sequence of described uniqueness can bound energy enough directly or indirectly in conjunction with unique polypeptide of the cap sequence in the exogenous RNA molecule of cracking.Described unique polypeptide also can be by composition coding of the present invention.In this embodiment, the exogenous RNA molecule of described unique polypeptide and cracking can produce the ring structure that can increase the translation efficiency of exogenous object protein in the exogenous RNA of the cracking molecule.For example, referring to Figure 13 B.
In other embodiments, composition of the present invention can comprise other polynucleotide sequence, this other polynucleotide sequence codified 3 ' end has can be in conjunction with the other RNA molecule of the nucleotide sequence in the sequence downstream that is positioned at cracking site upstream and encoding exogenous target protein.The expression of described other polynucleotide sequence can be by the promoters driven based on polymerase II.In this embodiment, described other RNA molecule can provide the poly-A tail of the translation efficiency of the exogenous object protein that can increase the exogenous RNA of autothermic cracking molecule in conjunction with the exogenous RNA molecule of cracking and for it.For example, referring to Figure 13 C.
3.3.
Can reduce the other structure of the translation efficiency of complete exogenous RNA molecule
The embodiment of the other structure that can reduce the complete translation efficiency of exogenous RNA molecule before it is cleaved has been described in this part.
In certain embodiments, described composition also can comprise can realize the exogenous RNA molecule directly or indirectly in the specific cleavage component that is positioned at the cracking that suppresses the sequence location downstream, and wherein said inhibition sequence is positioned at the cracking site downstream.Described specific cracking component for example can comprise:
(a) be positioned at the intramolecular specific nucleic acid sequence of described exogenous RNA, wherein said specific nucleic acid sequence can be selected from, but is not limited to: endonuclease recognition site, endogenous miRNA binding site, cis acting type ribozyme or miRNA sequence; Perhaps
(b) by the specific inhibitory RNA of composition of the present invention coding, wherein said specific inhibitory RNA can be selected from such as but not limited to the RNA of Microrna (miRNA), lasso trick form, short hairpin RNA (shRNA), siRNA expression structure territory, sense-rna, double-stranded RNA (dsRNA), siRNA (siRNA) or ribozyme.
In this embodiment, described specific cleavage component can be removed the poly A tract bar in the complete exogenous RNA molecule, to reduce the translation efficiency of exogenous object protein in the complete exogenous RNA molecule.For example, referring to Figure 13 D.
4.
Set forth and other embodiments
Term " enough complementarity " can include but not limited to can in conjunction with or complementary at least in part.In certain embodiments, the enough complementarity of term is in the scope of about 30-100%.For example, in certain embodiments, the enough complementarity of term are at least 30% complementarity.For example, in certain embodiments, the enough complementarity of term are at least 50% complementarity.For example, in certain embodiments, the enough complementarity of term are at least 70% complementarity.For example, in certain embodiments, the enough complementarity of term are at least 90% complementarity.For example, in certain embodiments, the enough complementarity of term are about 100% complementarity.
According to some embodiment, the cell that can insert/introduce composition of the present invention can be such as but not limited to: people's cell, zooblast, cultured cells, vegetable cell, primary cell, be present in the cell in the organism.
In certain embodiments, the specificity endogenous RNA molecule of cracking exogenous RNA molecule can be such as but not limited to: the peculiar Microrna of particular cell types, the peculiar miRNA of neoplastic cell, viral Microrna or similar RNA.The virus of viral miRNA of encoding can be selected from such as but not limited to double-stranded DNA virus, single-stranded DNA viruses, diplornavirus, diplornavirus, strand (normal chain) virus, strand (minus strand) virus or retrovirus.
In some exemplary, miRNAs is optional from such as, but not limited to miR-17-92 in the specificity of cracking exogenous RNA molecule, miR-221, miR-222, miR-146, miR-221, miR-21, miR-155, mir675, miR-10b, hsv1-miR-H1, hsv1-miR-H2, hsv1-miR-H3, hsv1-miR-H4, hsv1-miR-H5, hsv1-miR-H6, hsv2-miR-I, hcmv-miR-UL22A, hcmv-miR-UL36, hcmv-miR-UL70, hcmv-miR-UL112, hcmv-miR-UL148D, hcmv-miR-US4, hcmv-miR-US5-1, hcmv-miR-US5-2, hcmv-miR-US25-1, hcmv-miR-US25-2, hcmv-miR-US33, kshv-miR-K12-1, kshv-miR-K12-2, kshv-miR-K12-3, kshv-miR-K12-4, kshv-miR-K12-5, kshv-miR-K12-6, kshv-miR-K12-7, kshv-miR-K12-8, kshv-miR-K12-9, kshv-miR-K12-10a, kshv-miR-K12-10b, kshv-miR-K12-11, kshv-miR-K12-12, ebv-miR-BART1, ebv-miR-BART2, ebv-miR-BART3, ebv-miR-BART4, ebv-miR-BART5, ebv-miR-BART6, ebv-miR-BART7, ebv-miR-BART8, ebv-miR-BART9, ebv-miR-BART10, ebv-miR-BART11, ebv-miR-BART12, ebv-miR-BART13, ebv-miR-BART14, ebv-miR-BART15, ebv-miR-BART16, ebv-miR-BART17, ebv-miR-BART18, ebv-miR-BART19, ebv-miR-BART20, ebv-miR-BHRF1-1, ebv-miR-BHRF1-2, ebv-miR-BHRF1-3, bkv-miR-B1, jcv-miR-J1, hiv1-miR-H1, hiv1-miR-N367, hiv1-miR-TAR, sv40-miR-S1, MCPyV-miR-M1, hsv1-miR-LAT, hsv1-miR-LAT-ICP34.5, hsv2-miR-II, hsv2-miR-III, hcmv-miR-UL23, hcmv-miR-UL36-1, hcmv-miR-UL54-1, hcmv-miR-UL70-1, hcmv-miR-UL22A-1, hcmv-miR-UL112-1, hcmv-miR-UL148D-1, hcmv-miR-US4-1, hcmv-miR-US24, hcmv-miR-US33-1, hcmv-RNA β 2.7, ebv-miR-BART1-1, ebv-miR-BART1-2, ebv-miR-BART1-3, ebv-miR-BHFR1, ebv-miR-BHFR2, ebv-miR-BHFR3, hiv1-miR-TAR-5p, hiv1-miR-TAR-p, hiv1-HAAmiRNA, hiv1-VmiRNA1, hiv1-VmiRNA2, hiv1-VmiRNA3, hiv1-VmiRNA4, hiv1-VmiRNA5, hiv2-miR-TAR2-5p, hiv2-miR-TAR2-3p, mdv1-miR-M1, mdv1-miR-M2, mdv1-miR-M3, mdv1-miR-M4, mdv1-miR-M5, mdv1-miR-M6, mdv1-miR-M7, mdv1-miR-M8, mdv1-miR-M9, mdv1-miR-M10, mdv1-miR-M11, mdv1-miR-M12, mdv1-miR-M13, mdv2-miR-M14, mdv2-miR-M15, mdv2-miR-M16, mdv2-miR-M17, mdv2-miR-M18, mdv2-miR-M19, mdv2-miR-M20, mdv2-miR-M21, mdv2-miR-M22, mdv2-miR-M23, mdv2-miR-M24, mdv2-miR-M25, mdv2-miR-M26, mdv2-miR-M27, mdv2-miR-M28, mdv2-miR-M29, mdv2-miR-M30, mcmv-miR-M23-1, mcmv-miR-M23-2, mcmv-miR-M44-1, mcmv-miR-M55-1, mcmv-miR-M87-1, mcmv-miR-M95-1, mcmv-miR-m01-1, mcmv-miR-m01-2, mcmv-miR-m01-3, mcmv-miR-m01-4, mcmv-miR-m21-1, mcmv-miR-m22-1, mcmv-miR-m59-1, mcmv-miR-m59-2, mcmv-miR-m88-1, mcmv-miR-m107-1, mcmv-miR-m108-1, mcmv-miR-m108-2, rlcv-miR-rL1-1, rlcv-miR-rL1-2, rlcv-miR-rL1-3, rlcv-miR-rL1-4, rlcv-miR-rL1-5, rlcv-miR-rL1-6, rlcv-miR-rL1-7, rlcv-miR-rL1-8, rlcv-miR-rL1-9, rlcv-miR-rL1-10, rlcv-miR-rL1-11, rlcv-miR-rL1-12, rlcv-miR-rL1-13, rlcv-miR-rL1-14, rlcv-miR-rL1-15, rlcv-miR-rL1-16, rrv-miR-rR1-1, rrv-miR-rR1-2, rrv-miR-rR1-3, rrv-miR-rR1-4, rrv-miR-rR1-5, rrv-miR-rR1-6, rrv-miR-rR1-7, mghv-miR-M1-1, mghv-miR-M1-2, mghv-miR-M1-3, mghv-miR-M1-4, mghv-miR-M1-5, mghv-miR-M1-6, mghv-miR-M1-7, mghv-miR-M1-8, mghv-miR-M1-9 or sv40-miR-S1[34,35].The name of various miRNA molecules and sequence such as database http://www.mirbase.org/. are defined.
According to some embodiment, can be the protein of any kind by the exogenous object protein of exogenous RNA molecule encoding.For example, exogenous object protein can be selected from but be not limited to: alpha toxin, saporin, Zea mays RIP, barley RIP, wheat RIP, corn RIP, rye RIP, flax RIP, shiga toxin, will are congratulated sample RIP, momordin, thymidine kinase, Pokeweed antiviral protein, are spent more white tree toxalbumin, Pseudomonas exotoxin, ETA or its modified forms, ricin A chain, abrin A chain, diphtheria toxin Segment A or its modified forms, fluorescin, enzyme (such as for example, luciferase), structural protein or albuminoid.
In certain embodiments, described exogenous object protein can be the toxin that also can influence flanking cell.For example, described toxin can be selected from but be not limited to following complete form: Ricin, abrin, diphtheria toxin or its modified forms.In certain embodiments, described exogenous object protein can be enzyme for example, and the product of described enzyme also can kill flanking cell.This enzyme can be such as, but not limited to the HSV1 thymidine kinase.In certain embodiments, composition of the present invention also can comprise prodrug-ganciclovir, and ganciclovir is the substrate of HSV1 thymidine kinase.In some exemplary, described enzyme can be coli cytosine deaminase, and described composition also can comprise prodrug-5-flurocytosine (5-FC).
In certain embodiments, except the coding region of described exogenous object protein, the sequence of encoding exogenous target protein can comprise a plurality of introns of the expression that can increase target protein.In certain embodiments, intron can be the intron of a part for the natural gene of coding target protein.In certain embodiments, intron can be independent basis because of intron.In certain embodiments, the exogenous RNA molecule can be by any expression vector codes.For example, described exogenous RNA molecule can be the product that virus vector copies necessary gene by virus vector coding and described exogenous object protein, and wherein said virus vector copies and kill described cell in response to the existence of miRNAs in the cell internal specific in reproduction process.Virus vector also can comprise, and for example, can stop the gene that virus vector copies when specific molecular (for example, TetR-VP16/ Vibravenos) is present in the cell.Wherein when inferring the sudden change that virus vector has obtained to be enough to copy in the cell that does not comprise miRNAs in the specificity, can use that described specific molecular copies with all virus vector in the cessative aspect and then after in vivo most of virus vector degraded, can use new virus vector again.Virus vector also for example can comprise, when (for example having specific prodrug, thymidine kinase/ganciclovir) gene that can cell killing the time, wherein when inferring the sudden change that virus vector has obtained to be enough to copy in the cell that does not comprise miRNAs in the specificity, can use this specific prodrug to kill all virus vector in the body and can use new virus vector again then.
In some exemplary, the exogenous RNA molecule can be by the virus vector coding that can copy in the mode of cell killing in reproduction process.In this embodiment, miRNAs is not present in patient's the target cell (for example, cancer cells) in the specificity, but most of normal cells or non-metastatic that the interior miRNAs of this specificity is present in the patient cause in the tumour cell.In this example, exogenous object protein is that toxin is for example such as ricin A chain, abrin A chain, diphtheria toxin Segment A or its modified forms.Cause tumour cell it kills this cell when virus vector enters normal cell or non-metastatic, and when virus vector enters target cell (cancer cells), it is kill cancer cell in the virus vector reproduction process, so the big concentration of virus vector is present in tumor region.This virus vector also can comprise, can stop the gene that virus vector copies when specific molecular (for example, TetR-VP16/ Vibravenos) is present in the cell.Wherein when inferring that virus vector has obtained to be enough to comprising the sudden change that copies in the cell of miRNAs in the specificity, can use described specific molecular copying and then after the degraded of the most of virus vector in somatocyte, can use new virus vector again with all virus vector in the cessative aspect.This virus vector can comprise, when (for example having specific prodrug, thymidine kinase/ganciclovir) gene that can cell killing the time, wherein when inferring that virus vector has obtained to be enough to comprising the sudden change that copies in the cell of miRNAs in the specificity, can use this specific prodrug to kill all virus vector in the body and can use new virus vector again then.
According to some embodiment, suppress sequence and can be in case with after the sequence of encoding exogenous target protein is separated, then the sequence that can be expressed of described exogenous object protein or the part of this sequence.When suppressing sequence and do not separate with the sequence of encoding exogenous target protein, when it was in its specific background in the exogenous RNA molecule, it can suppress expression of exogenous object protein.Suppress the part that is arranged in its specific environment that sequence also can include only above-mentioned any inhibition sequence.For example, be respectively--UG-3 ' or--under the background of G-3 ', suppressing sequence can be independent A or 5 '-AU-3 ' part, rather than do not meet reading frame (out of reading frame) 5 '-AUG-3 ' the inhibition sequence (just, the exogenous RNA molecule 5 ' end comprises and do not meet 5 ' of reading frame-AUG-3 ', but the sequence of separating is 5 '-AU-3 ' part).
In another embodiment of the invention, composition of the present invention also can comprise the polynucleotide sequence that coding can suppress the particular functionality RNA of endogenous exonuclease expression directly or indirectly.Described particular functionality RNA can be such as but not limited to: the RNA of Microrna (miRNA), lasso trick form, short hairpin RNA (shRNA), siRNA expression structure territory, sense-rna, double-stranded RNA (dsRNA), siRNA (siRNA) or ribozyme.
In another embodiment of the invention, above-mentioned binding site can be a plurality of binding sites at identical or different miRNA, makes wherein said " cracking site upstream " also comprise " all cracking site upstreams ".Similarly, wherein said " cracking site downstream " also comprises " all cracking site downstreams ".In certain embodiments, when described a plurality of binding sites during at different endogenous miRNA, even only there is a kind of miRNA in the cell, exogenous object protein can be expressed.For example, referring to Figure 14 A, 14B, 14C, 14D.
In certain embodiments of the invention, the exogenous RNA molecule also can comprise the one or more other binding site at miRNAs in the specificity, and wherein said one or more other binding sites have to disturb by RNA makes that miRNAs instructs the exogenous RNA molecule in enough complementarity of the cracking site place cracking of uniqueness in the specificity.Each unique cracking site can be positioned at the sequence upstream that each other binding site and cracking site that each is unique can be positioned at the described exogenous object protein of coding.The exogenous RNA molecule also can comprise one or more initiator codons of all unique cracking site upstreams, and wherein the sequence of each initiator codon and encoding exogenous target protein is not in same reading frame.For example, initiator codon can be mainly by 5 '-AUG-3 ' forms, wherein at least one initiator codon is positioned at Kozak consensus sequence or any other translation initiation element.Initiator codon can be TISU element [38] for example.According to some embodiment, after described composition being incorporated in the cell that comprises miRNAs in the specificity, the exogenous RNA molecule can be transcribed and by miRNAs in the described specificity in cracking site and each unique cracking site place cracking, make the sequence of encoding exogenous target protein separate with the inhibition sequence, and separate with each initiator codon, and exogenous object protein can be expressed.For example, referring to Figure 15 A.
In certain embodiments, composition of the present invention also can comprise can realize being positioned at the exogenous RNA molecule directly or indirectly in the cracking component of the cracking of the position of each upstream from start codon, and wherein said cracking component for example is:
(a) be positioned at the intramolecular nucleotide sequence of described exogenous RNA, wherein said nucleotide sequence is: endonuclease recognition site, endogenous miRNA binding site, cis acting type ribozyme or miRNA sequence; Perhaps
(b) by the inhibitory RNA of described composition coding, wherein said inhibitory RNA is: the RNA of Microrna (miRNA), lasso trick form, short hairpin RNA (shRNA), siRNA expression structure territory, sense-rna, double-stranded RNA (dsRNA), siRNA (siRNA) or ribozyme.For example, referring to Figure 15 B.
According to some embodiment, composition of the present invention can comprise one or more polynucleotide molecules, such as dna molecular for example, RNA molecule or the two.In one embodiment, described composition can comprise when only having miRNAs molecule in the specificity in cell at the dna molecular of cell inner expression exogenous object protein, and miRNAs can be the miRNA of for example cell, viral miRNA or similar miRNA in the wherein said specificity.Described dna molecular can comprise the polynucleotide sequence of encoding exogenous RNA molecule, described exogenous RNA molecule is to comprise following RNA molecule: the sequence of encoding exogenous target protein, the binding site at miRNAs in the specificity of the upstream of the sequence of encoding exogenous target protein, the other binding site at miRNAs in the specificity in the downstream of the sequence of encoding exogenous target protein, at least two are suppressed that sequence-is positioned at 5 of described exogenous RNA molecule ' end and another is positioned at 3 of described exogenous RNA molecule ' end, and wherein each inhibition sequence can suppress expression of exogenous object protein.Therefore, when only having in the specificity miRNAs in cell, described two are suppressed sequences and can separate with the sequence of encoding exogenous target protein, and exogenous object protein can be at cell inner expression.These suppress sequences can be any in the above-mentioned sequence.For example, referring to Figure 15 C.
According to other embodiments, described composition can comprise the dna molecular of expressing exogenous object protein at described cell when only having miRNAs in two specific specificity in cell.Described dna molecular can comprise the polynucleotide sequence of encoding exogenous RNA molecule, and described exogenous RNA molecule is to comprise following RNA molecule: the binding site at miRNAs in first specificity of the sequence upstream of the sequence of encoding exogenous target protein, encoding exogenous target protein, the sequence downstream of encoding exogenous target protein suppress that sequence-is positioned at 5 of described exogenous RNA molecule ' end and another is positioned at 3 of described exogenous RNA molecule ' end at another binding site of miRNAs in second specificity and at least two.Each suppresses sequence can suppress expression of exogenous object protein, make when in existence two specific specificity in the cell during miRNAs, described two inhibition sequences can be separated with the sequence of encoding exogenous target protein, and exogenous object protein can be at cell inner expression.These suppress sequences can be any in the above-mentioned sequence.For example, referring to Figure 15 D.
According to other embodiments, when when only there is multiple different miRNA simultaneously in needs in cell, just expressing exogenous object protein, the composition of the present invention multiple exogenous RNA molecule that can comprise or encode, wherein the structure of each exogenous RNA molecule can be aforesaid.Described exogenous RNA molecule can be similar or different.In these exogenous RNA molecules each can comprise the different sequences of the different miRNA binding sites target protein different with coding, and wherein all these different target proteins can produce new function together in cell.For example; when described multiple different miRNA comprises three kinds of different miRNA; three kinds of different target proteins by three kinds of different exogenous RNA developed by molecule can be selected from: protective antigen (PA), edema factor (EF) and lethal gene (LF); wherein when having described three kinds of different miRNA in the cell simultaneously, these 3 kinds of albumen: the anthrax toxin that can cause necrocytosis is expressed and produced together to protective antigen (PA), edema factor (EF) and lethal gene (LF).
In another embodiment, the exogenous RNA molecule also can have the RNA signal for locating for Subcellular Localization (comprise common translation input) between the sequence of cracking site and encoding exogenous target protein, wherein suppressing sequence, can to suppress Subcellular Localization for the function of the RNA signal for locating of Subcellular Localization and wherein said exogenous RNA molecule be that the correct expression of exogenous object protein is necessary.For example, referring to Figure 16 A, 16B.
In other embodiments, suppress the initiator codon that sequence can comprise the cracking site upstream, wherein said initiator codon mainly by 5 '-AUG-3 ' forms.Suppress the nucleotide sequence that sequence also can comprise the aminoacid sequence in coding next-door neighbour initiator codon downstream, the sequence of wherein said nucleotide sequence and encoding exogenous target protein is in the same reading frame.Described aminoacid sequence can suppress the function for the sorting signals of the Subcellular Localization of exogenous object protein, and wherein the Subcellular Localization of exogenous object protein is that its correct expression is necessary, for example, and referring to Figure 16 C.
In another embodiment of the invention, the exogenous RNA molecule does not comprise the terminator codon in the sequence downstream of encoding exogenous target protein.Described inhibition sequence can be positioned at the sequence downstream of encoding exogenous target protein, and the sequence of wherein said inhibition sequence and encoding exogenous target protein is in same reading frame, and described inhibition sequence encoding is selected from the aminoacid sequence by the following group of forming:
(a) can suppress the aminoacid sequence of the function of exogenous object protein;
(b) be the aminoacid sequence that is used for the sorting signals of Subcellular Localization;
(c) be the aminoacid sequence of proteolytic degradation signal;
(d) can suppress aminoacid sequence for the function of the sorting signals of the Subcellular Localization of exogenous object protein; With
(e) can suppress by the aminoacid sequence in the cracking of the nucleotide sequence coded peptide sequence between the starting codon of the sequence of cracking site and encoding exogenous target protein, the sequence of wherein said nucleotide sequence and encoding exogenous target protein is in the same reading frame and wherein said peptide sequence can be by the protease cracking in the mammalian cell.(also be reported in people's cell, during the mRNA that translates the brachymemma that does not contain terminator codon, rrna be parked in the terminator codon place and relevant tRNA molecule still maintenance be combined with polypeptide chain and rrna, but, in the centre of translation process, peptidyl-tRNA material is possible [32] by the processing of endoplasmic reticulum signal peptidase).For example, referring to Figure 16 D.
5.
Synthesizing of composition of the present invention
According to some embodiment, and describe in detail as mentioned, described composition can comprise and comprise or one or more polynucleotide molecules of encoding exogenous RNA molecule.Described polynucleotide molecule can be one or more dna moleculars, one or more RNA molecules or its combination.In some exemplary, described composition can comprise one or more dna moleculars of encoding exogenous RNA molecule.The dna molecular of encoding exogenous RNA molecule can be become also can provide copying on a large scale of DNA and comprise the multiple host carrier system/construct of transcribing necessary element that instructs the exogenous RNA molecule by recombination to construct.These carriers are introduced cause transcribing of capacity exogenous RNA molecule in the cell in target cells.For example, carrier can be introduced and make it be absorbed by cell in the body to and guide transcribing of exogenous RNA molecule.This carrier can remain free or be colored body and integrate, as long as it can transcribe the exogenous RNA molecule that produces expectation.These carriers can make up by recombinant DNA technology method well known in the art maybe can be by any method preparation for the synthesis of dna molecular known in the art.
According to some embodiment, the recombinant DNA construction body of encoding exogenous RNA molecule can comprise plasmid for example, cosmid, virus vector or known in the art for the target cell of expectation (such as, for example mammalian cell (for example, people's cell, mouse cell), birds cell, vegetable cell and similar cell) in any other carrier of copying and expressing.The expression of exogenous RNA molecule can be regulated by any promotor that plays a role in the target cell of expectation known in the art.These promotors can be inducibility or composition.These promotors comprise, such as but not limited to: the promotor that comprises in 3 of SV40 early promoter zone, Rous sarcoma virus ' terminal repetition district of length, the promotor of herpes thymidine kinase, the regulating and controlling sequence of metallothionein gene, viral CMV promotor, human chorionic gonadotropin-β promotor etc.In certain embodiments, promotor can be promotor (that is, the promotor of being identified by RNA Pol.I) such as, the promotor of rDNA (rDNA) gene for example of rna plymerase i.In these embodiments, the termination signal of external source purpose RNA molecule can be RNA Pol.I termination signal or rna plymerase ii termination signal (such as for example, the poly a-signal).The plasmid of any kind, cosmid, YAC or virus vector can be used to prepare the recombinant DNA construction body that can be introduced directly into target cell/cell mass or tissue site.Selectively, can use the virus vector that optionally infects the target cell of expectation.
According to some embodiment, for the formation of the transgenic organism of resisting virus infection or cancer, the carrier of expectation encoding exogenous RNA molecule will have selection marker.Can use several selective systems, include but not limited to the selection of the expression in tk-, hgprt-or aprt-deficient cell respectively of herpes simplex virus thymidine kinase, xanthoglobulin-guanine phosphoribosyltransferase and adenine phosphoribosyl transferase protein.In addition, the metabolic antagonist resistance dihydrofolic acid transferring enzyme (dhfr) that provides the resistance of methotrexate is provided, xanthine-guanine phosphoribosyl transferase (gpt) to the resistance of mycophenolic acid is provided, provides the Xin Meisu (neo) of the resistance of aminoglycoside G-418 and basis to the selection of the hygromycin B phosphotransferase (hygro) of the resistance of Totomycin is provided.
According to some embodiment, the carrier that is used for practice of the present invention can be any expression vector.In some exemplary, the exogenous RNA molecule is encoded by virus expression carrier.Virus expression carrier can be selected from, but is not limited to: herpetoviridae (Herpesviridae), Poxviridae (Poxyiridae), Adenoviridae (Adenoviridae), Papillomaviridae (Papillomaviridae), Parvoviridae (Parvoviridae), Hepadnaviridae (Hepadnoviridae), Retroviridae (Retroviridae), Reoviridae (Reoviridae), Filoviridae (Filoviridae), Paramyxoviridae (Paramyxoviridae), Pneumovirinae section (Pneumoviridae), Rhabdoviridae (Rhabdoviridae), orthomyxoviridae family (Orthomyxoviridae), Bunyaviridae (Bunyaviridae), Hantaan virus section (Hantaviridae), Picornaviridae (Picornaviridae), Caliciviridae (Caliciviridae), Togaviridae (Togaviridae), flaviviridae (Flaviviridae), Arenaviridae (Arenaviridae), coronaviridae (Coronaviridae), or Hepacivirus (Hepaciviridae).Virus expression carrier can include but not limited to that also its cytotaxis is exposed to the fibrinous adenovirus terminal knot shape structural domain (HI ring) of fiber surface and reformed adenovirus carrier by replacement.
In certain embodiments, composition of the present invention can comprise one or more RNA molecules, and described one or more RNA molecules can be for example strand or the double-stranded or derivatives thereof of endogenous RNA molecule own or modified forms.The exogenous RNA molecule can have such as, but not limited to following Nucleotide: the key of deoxyribonucleotide, ribonucleotide, phosphodiester bond, modification or the base except five kinds of bases (VITAMIN B4, guanine, thymus pyrimidine, cytosine(Cyt) and uridylic) that biologically exist.
According to some embodiment, the exogenous RNA molecule can be by any method preparation for the synthesis of the RNA molecule known in the art.For example, the exogenous RNA molecule can use commercially available reagent and synthesizer by method chemosynthesis well known in the art.Selectively, the exogenous RNA molecule can produce by the dna sequence dna of transcribing encoding exogenous RNA molecule in external and the body.These dna sequence dnas can be integrated into has integrated suitable R NA polymerase promoter for example in the various carriers of T7 or SP6 polymerase promoter.The exogenous RNA molecule can for example produce on SPS65 in-vitro transcription high yield ground by using plasmid.In addition, the RNA amplification method for example Q-β amplification can be used to produce the exogenous RNA molecule.
In certain embodiments, the dna molecular of exogenous RNA molecule or encoding exogenous RNA molecule can be modified at for example base portion, sugar moieties or phosphoric acid skeleton place, with the stability of improving molecule, hybridization, to intracellular transportation and similar feature.In addition, can modify to reduce susceptibility to nuclease degradation.The dna molecular of exogenous RNA molecule or this exogenous RNA molecule of encoding can have for example peptide (for example, be used for body in target host cell receptor) or help to pass the agent of cytolemma or hemato encephalic barrier transportation, cracking agent or the intercalator that hybridization triggers of other side groups.Can introduce the multiple modification that other are known as increasing cell inner stablity and the means of transformation period.Possible modification includes but not limited to, to 5 of molecule ' and/or 3 ' end add the flanking sequence of ribonucleotide or deoxyribonucleotide.Under the certain situation of the stability that expectation therein increases, can preferably have between the Nucleotide of modification key for example 2 '-the methylated nucleic acid of O-.The nucleic acid that comprises key between the Nucleotide of modification can use reagent well known in the art and method synthetic.
According to other embodiments, the dna molecular of exogenous RNA molecule or encoding exogenous RNA molecule can be by any suitable means as known in the art (such as, for example reverse-phase chromatography or gel electrophoresis) purifying.
In certain embodiments, the cell of the virus vector of generation encoding exogenous RNA also can be used for being transplanted in patient's body with continued treatment.These cell portabilities cause its dead specific gene in the time of can having specific molecular (for example, HSV1 thymidine kinase/ganciclovir) in blood.
In certain embodiments, the exogenous RNA molecule can be RNA molecule or replicability RNA molecule.Replicability RNA molecule is the RNA molecule that comprises with the sequence of exogenous RNA complementary element, and wherein said replicability RNA molecule can be in time multiplexed cell system to form described exogenous RNA molecule.
6.
The purposes of composition of the present invention and using
According to some embodiment, composition of the present invention can have multiple different application and for example comprise, but be not limited to: regulate gene expression, targeted cells death, treat various illnesss and illness such as for example: treatment hyperplasia illness is cancer, treatment transmissible disease HIV for example for example, forms transgenic organism, suicide gene therapy and similarly uses.Described composition can be used on the different organisms, such as for example, and Mammals (for example people, mouse), birds, plant and similar organism.Described composition can be used on different cells (cultivation and/or body in), on the health of tissue, organ and/or organism.
In certain embodiments, composition of the present invention can be used to express in the cell of miRNAs in being expressed as the specificity of viral miRNA and/or the activation virulent gene, to kill the cancer cells of expressing this virus miRNA or to kill the cell of virus infection.In another embodiment of the invention, composition of the present invention can be used to express and/or the activation virulent gene in as the cell of specificity endogenous virus miRNA comprising carinogenicity miRNA (miRNA that is raised strongly) in cancer cells, to kill these cells.
In certain embodiments, composition of the present invention can be used to express in the presence of viral miRNA or carinogenicity miRNA and/or activate reporter gene to diagnose the illness as virus infection or cancer.In another embodiment, transfection stably the cell of carrier of encoding exogenous RNA molecule can be used for forming the transgenic organism of opposing virus infection or cancer.In another embodiment, composition of the present invention can be used to transfectional cell stably, and described cell is used to form and can activates reporter gene with the transgenic organism of diagnosis disease of viral infection in the presence of viral miRNA.In another embodiment, composition of the present invention can be used to monitor in real time the function of miRNA in the cell also for diagnosing the disease (for example, cancer and virus infection) that relates to the interior miRNA of formation of cell or miRNA rise.
According to some embodiment, multiple delivery system is known and can be used to composition of the present invention is transferred in the cell, such as for example, be encapsulated in liposome, particulate, in the microcapsule, can express the reconstitution cell of described composition, receptor-mediated endocytosis, composition of the present invention is configured to the part of virus vector or other carriers, can be replicated and in reproduction process cell killing and comprise the virus vector of composition of the present invention not, reproducible and comprise the virus vector of composition of the present invention not, injection produces the cell of the virus vector that comprises composition of the present invention, injection DNA, electroporation, transfection and the similar approach of calcium phosphate mediation, any other method leaved for development known in the art or following.
, and do not expect by theory or mechanism constraint ground that the compositions and methods of the invention can provide " whole or nothing (all or none) " reaction of specific and target according to some embodiment in cell.In other words, the compositions and methods of the invention make the exogenous RNA molecule only in comprising specificity cleaved in the target cell of miRNAs (and therefore, express and the activation exogenous object protein), and do not comprise that the cell of described endogenous miRNA can not be acted on by composition of the present invention.Therefore the compositions and methods of the invention can provide security and the control of increase, do not express (leakiness of expression) because observe the omission of exogenous object protein in the cell that does not comprise described endogenous miRNA.
According to some embodiment, provide to be used for the method for killing the specific cells group, wherein said cell mass comprises uniqueness and to miRNAs in the specificity of these cell-specifics; Described method comprises introduces cell with composition of the present invention, wherein said composition comprises for one or more polynucleotide that only the miRNAs cell instructs exogenous object protein to express in expression specificity, and wherein said one or more polynucleotide comprise or encode and comprise following exogenous RNA molecule: the sequence of encoding exogenous target protein; The inhibition sequence that can suppress expression of exogenous object protein; And at the binding site of miRNAs in the described specificity.
According to some embodiment, described exogenous object protein can be the protein that can destroy cell function and therefore cause any kind of necrocytosis.Described protein can be selected from such as, but be not limited to the protein of following type: toxin, cytostatic agent, cell growth regulator, cell signal pathway inhibitor, cell signal pathway modulators, cell permeability modulators, cell processes conditioning agent (modulators of cellular processes) and similar protein.
According to some embodiment, carrier is provided, such as for example expression vector (virus vector or non-virus carrier), described expression vector comprises one or more polynucleotide sequences of encoding exogenous RNA molecule, and wherein said exogenous RNA molecule comprises the sequence of encoding exogenous target protein; Can suppress the inhibition sequence that described exogenous object protein is expressed; And at the binding site of miRNAs in the specificity.Have when described carrier being incorporated in the cell that comprises miRNAs in the specificity at certain sequence in the miRNAs in the binding site of miRNAs in the described specificity and this specificity, make that miRNAs instructs described exogenous RNA molecule in enough complementarity of cracking site cracking in the described specificity.Described cracking site can be positioned at the binding site at the specificity miRNAs, and in addition, described cracking site is between the sequence that suppresses sequence and the described exogenous object protein of encoding.In certain embodiments, described one or more polynucleotide sequences are dna sequence dnas.In certain embodiments, described one or more polynucleotide sequences are RNA sequences.As known in the art, carrier also can comprise this carrier needed multiple other polynucleotide sequences of operation (such as, for example, regulating and controlling sequence, non-coding sequence, structure sequence and similar sequence).
According to other embodiments, the present invention also provides the composition of the present invention that comprises significant quantity and the pharmaceutical composition of pharmaceutically acceptable vehicle (carrier).Term " pharmaceutically acceptable " refers to be listed in by the regulator of federation or state government approval or in American Pharmacopeia or other pharmacopeia of generally acknowledging for animal and people more particularly.Term " vehicle " refers to thinner, adjuvant, vehicle or the vehicle used with therapeutical agent.
According to some embodiment, described pharmaceutical composition can be by known any route of administration, such as such as, but not limited to: the curee to needs uses through intestines, parenteral, injection, part and similar approach.In certain embodiments, can expect the target region of needs treatment is used pharmaceutical composition of the present invention partly.This can be by for example and be not limited to: perioperative local infusion, topical application are (for example, be combined with postoperative wound dressings), by injection, by conduit, by suppository or realize by implant, described implant is porose, atresia or gelatinous material, comprise for example silicone rubber membrane (sialastic membranes) of film, or fiber.Topical application also can be passed through controlled release-drug delivery system, for example for example controlled release polymer or hydrogel realization of nano particle, matrix.
In certain embodiments, composition of the present invention can be used with the amount that produces expectation function in target cell/tissue effectively.The effective dose of composition of the present invention can be by answer well known to those skilled in the art such as biological half-life, bioavailability and toxicity the program of parameter determine.The significant quantity of composition of the present invention depends on the character of the disease of being treated or illness and can determine by standard clinical techniques.In addition, external test is optionally with helping determine the optimal dose scope.Application process also can include but not limited to patient's blood flow is injected composition of the present invention lastingly or continuously.
In certain embodiments, can be to multiple organism such as for example, Mammals, birds, plant and similar organism applying said compositions and comprise the pharmaceutical composition of described composition.For example, can be to humans and animals applying said compositions and the pharmaceutical composition that comprises described composition.
In other embodiments, the present invention also provides cartridge bag or the medicine box of one or more containers that comprise one or more compositions that are filled with pharmaceutical composition of the present invention, what be connected alternatively with described container can be that this bulletin has reflected the permission of the manufacturing, use or the sale that obtain mechanism and use at the human or animal by the bulletin of the form of government organs' regulation of manufacturing, use or the sale of supervision medicine or biological products.
Embodiment
The following example is by exemplary and provide and be the embodiment of optimum implementation of the present invention without limitation.
Embodiment 1-is specific expressed by the exogenous object protein of exogenous RNA coding
The routine operation method of the experiment of describing among the embodiment 1:
In transfection the day before yesterday, with about 120,000 the T293 cell inoculations in every hole in 24 orifice plates, transfection same day with each hole of following cotransfection:
1. sea pansy (renila)/luciferase plasmid-170ng expresses sea pansy Ying Guangsumeijiyin ﹠amp; The plasmid of Lampyridea luciferase genes (plasmid E11, Psv40-intron-MCS-RLuc---Phsvtk-Fluc, SEQ ID NO:22, or plasmid E65, Psv40-intron-Tsp-TD1-TLacZ-RLuc-PTS-60ATG---Phsvtk-Fluc, SEQ ID NO.23).
2. the tested tested plasmid (as will be detailed later) of plasmid=30ng.
3.siRNA+ or the siRNA-=10 picomole can be induced by the siRNA duplex molecule (siRNA+) of tested plasmid-encoded mRNA cracking or not induced siRNA duplex molecule (siRNA-) by tested plasmid-encoded mRNA cracking.(described below).
Use lipofectamine2000 transfection reagent (Invitrogen) to carry out transfection according to producer's explanation.After the transfection 48 hours, use dual luciferase report to measure the expression of test kit (Promega) and photometer (glomax20/20promega) measurement sea pansy luciferase genes, and definite relative light unit (RLU).
Tested plasmid can be the following plasmid of any kind:
Negative controlThe plasmid of=diphtheria toxin (DTA) that do not encode;
Positive controlThe plasmid of=composition ground coding diphtheria toxin (DTA);
Tried plasmidThe plasmid of=composition of the present invention namely comprises the plasmid at the target site of siRNA+ between the downstream sequence that suppresses sequence and coding diphtheria toxin (DTA).For being tried plasmid, when the siRNA+ of cotransfection cracking is tried the inhibition sequence of plasmid, thereby the cell-expression of minimizing sea pansy luciferase and total RLU observed value of expressing diphtheria toxin can be expressed and be killed to diphtheria toxin.
Also test tested plasmid respectively with 2 kinds of different siRNA-with 2 kinds of different siRNA+, and triplicate separately.
The following calculating of result:
The activation multiple=in 2 kinds of siRNA-each and tried plasmid in the presence of RLU (relative light unit) mean value (6 hole) measured divided by the RLU mean value (3 hole) that uses a kind of among the siRNA+ and tried plasmid.
Omit multiple=use all siRNA-/+and the RLU mean value of negative control plasmid divided by using each among 2 kinds of siRNA-and being tried the RLU mean value of plasmid.
SiRNA+/-RLU=in the presence of a kind of siRNA+ of cotransfection or the siRNA-of two kinds of cotransfections in the presence of the RLU mean value measured independently.
Use the common and known method of using in the biology field to make up these plasmids.The skeleton carrier that is used for this paper structure plasmid described below is: psiCHECK
TM-2 carriers (promega, catalog number (Cat.No.) C8021) or pcmv6-A-GFP (OriGene, catalog number (Cat.No.) PS100026).Be described in further detail as hereinafter trying plasmid about these, the additional title of each plasmid is represented the sequence that comprises in this plasmid sequence.
The siRNA sequence:
1.RL duplex (Dharmacon, catalog number (Cat.No.) P-002070-01-20) (SEQ ID NO.65 (sense strand) and SEQ ID66 (antisense strand)).
2.GFP duplex II (Dharmacon, catalog number (Cat.No.) P-002048-02-20), (SEQ ID NO.67 (sense strand) and SEQ ID NO.68 (antisense strand)).
3.siRNA-contrast (Sigma, catalog number (Cat.No.) VC30002000010), (SEQ ID NO.69 (sense strand) and SEQ ID NO.70 (antisense strand)).
4. anti-β Gal siRNA-1 ((target site: Tlacz (SEQ ID NO.71)), Dharmacon, catalog number (Cat.No.) P-002070-01-20) (SEQ ID NO.72 (sense strand) and SEQ ID NO.73 (antisense strand)).
5. luciferase GL3 duplex ((target site: Tfluc (SEQ ID NO.74)), Dharmacon, catalog number (Cat.No.) D-001400-01-20), (SEQ ID NO.75 (sense strand) and SEQ ID NO.76 (antisense strand)).
6.GFP duplex I ((target site: TD1, (SEQ ID NO.77)), Dharmacon, catalog number (Cat.No.) P-002048-01-20), (SEQ ID NO.78 (sense strand) and SEQ ID NO.79 (antisense strand)).
(7.TCTL (target site: TCTL (SEQ ID NO.80)), SEQ ID NO.81 (sense strand) and SEQ ID NO.82 (antisense strand)).
In each experiment, have the siRNA that is tried the target site in the plasmid and be used as siRNA+, and do not have other siRNA of target site corresponding in the tested plasmid to be used as siRNA-.
The negative control plasmid:
1.E34(SEQ?ID?NO.10)-Pcmv-4ORF
∧-
TD1-Tfluc---Psv40-TGFP。
(2.E71 SEQ ID.NO.17)-Psv40-intron-4ORF
∧---Phsvtk-Fluc.
3.E38-3CARz-4S&L。Between pacI and XhoI restriction site, inset E38 (SEQ ID.NO.19) is connected in the PMK shuttle vectors (GeneArt).
The positive control plasmid:
1.E28(SEQ?ID.NO.11)-Pcmv-
Tfluc-TD1-cDTAWT---Psv40-TGFP.
2.E20(SEQ?ID.NO.12)-Pcmv-nsDTA---Psv40-TGFP
(3.E70 SEQ ID.NO.13)-Psv40-intron-cDTAWT---Phsvtk-Fluc
4.E3(SEQ?ID.NO.14)-Pcmv-KDTA---Psv40-TGFP
5.E89(SEQ?ID.NO.15)-Pcmv---DT
∧A---Psv40-TGFP
6.E110(SEQ?ID.NO.16)-Pcmv-D5
∧TA---Psv40-TGFP
7.E4(SEQ?ID.NO.18)-Pcmv-KDTA---Psv40-Hygro
8.E10(SEQ?ID.NO.20)-Pef1-DTA24---ZEO::GFP-Pcmv
(9.E143 SEQ ID.NO.21)-3 poly A-Prp119-cDTAWT---Phsvtk-Fluc
Tried plasmid
(1.E80 SEQ ID.NO.1)-Pcmv-4ORF
∧-TD1-Tfluc-S-cDTAWT---Psv40-TGFP (pCMV promotor (nt.420-938 of SEQ ID NO.1); 4ORF
∧Among=4 continuous ORF by the following inhibition sequence of forming: 9 TISU sequences and 57 kozak sequences are 57,57,36,36,21,21,21 and 21nt (nt1027-3547 of SEQ ID NO.1) between the adjacent ATG codon.The one ORF (nt.1031-1651 of SEQ ID NO.1) is 621nt﹠amp; From TISU (nt.1027-1038 of SEQ ID NO.1) translation, and ensuing 3 ORF
∧(nt.1662-2996, nt.2306-2941 and the nt2951-3547 of SEQ ID NO.1) translates from the Kozak sequence.(cDTAwt=does not contain promotor/montage/termination/poly A site and contains the wt DTA encoding sequence of Kozak sequence (nt3568-4155 of SEQ ID NO.1) and stops before last ORF (nt2951-3547 of SEQ ID NO.1) at the encoding sequence of wild-type DTA; Next be the TGFP encoding sequence under the control of SV40 promotor.This plasmid also comprises target site TD1 (SEQ ID NO.77) and Tfluc (SEQ ID NO.74).
(2.E54 SEQ ID.NO.2)-Pcmv-4CARZ-PTS-60ATG
∧-3ORF
∧-TD1-Tfluc-incDTAWT---Psv40-TGFP (pCMV promotor (Nucleotide (nt.) 420-938 of SEQ ID NO.2); 4CAR=4 cis acting ribozyme (nt.1013-1373 of SEQ ID NO.2); PTS=peroxysome target signal (nt.1420-1500 of SEQ ID NO.2); 60ATG
∧=61ATG, 46 in the Kozak sequence, almost between per two ATG every 53nt (nt.1534-4554 of SEQ ID NO.2), and terminator codon is in DTA encoding sequence (nt.6745-7332 of SEQ ID NO.2); TGFP encoding sequence (nt.8452-9143 of SEQ ID NO.2) under psv40 promotor (nt.8092-8399 of the SEQ ID.NO.2) control.This plasmid also comprises target site TD1 (SEQ ID NO.77) and Tfluc (SEQ ID NO.74).
(3.E113 SEQ ID.NO.3)-Pcmv-4ORF
∧-TD1-Tfluc-PK-D5
∧TA---Psv40-TGFP (pCMV promotor (nt.420-938 of SEQ ID NO.3); 4ORF
∧(nt.1027-3547 of SEQ ID NO.3); PK=false knot (pseudoknot)-stem and ring, the wherein hybridization of starting codon of Huan 6nt and DTA (nt3561-3611 of SEQ ID No.3); 5
∧=5 encoding sequences that are positioned at DTA (nt.3609-3806 of SEQ ID NO.3 and comprise people's intron (nt.3712-3801,3856-3960,4066-4173,4380-4519 and the 4617-4783 of SEQ ID NO.3) be used to the sequence that is rich in T of the Transcription Termination that makes RNA polymerase 1 and/or 3, these introns are embedded in the cDTAwt encoding sequence; TGFP encoding sequence (nt5906-6597 of SEQ ID NO.3) under psv40 promotor (nt.5546-5853 of the SEQ ID NO.3) control.This plasmid also comprises target site TD1 (SEQ ID NO.77) and Tfluc (SEQ ID NO.74).
(4.E91 SEQ ID.NO.4)-Pcmv-4ORF
∧-TD1-Tfluc-DT
∧A---Psv40-TGFP (pCMV promotor (nt.420-938 of SEQ ID NO.4), 4ORF
∧(nt.1027-3507 of SEQ ID NO.4); DT
∧A=contains from the intron of human collagen 16A1 gene and does not contain the kozak DTA (nt.3520-4444 of SEQ ID NO.4) of promotor/montage/poly a-signal; TGFP encoding sequence (nt.5544-6235 of SEQ ID NO.4) under pSV40 promotor (nt.5184-5491) control.This plasmid also comprises target site TD1 (SEQ ID NO.77) and Tfluc (SEQ ID NO.74).
(5.E112 SEQ ID.NO.5)-Pcmv-4ORF
∧-2xTLacZin intron-8X[TCTL+TD1]-PK-D5
∧TA---Psv40-TGFP (pCMV promotor (nt.420-938 of SEQ ID NO.5), 4ORF
∧(nt.1027-3436 of SEQ ID NO.5); 2 TLacZ targets in the intron of 2xTLacZin intron=commercially available plasmid pSELECT-GFPzeo-LacZ (nt.3438-3638 of SEQ ID NO.5); 8X[TCTL+TD1] (nt.3647-4052 of SEQ ID NO.5); PK=false knot-stem and ring, the wherein hybridization of starting codon of Huan 6nt and DTA (nt4059-4109 of SEQ ID NO.5); 5
∧=5 encoding sequences that are positioned at DTA (nt.4107-5304 of SEQ ID NO.5 and comprise people's intron (nt.4210-4299,4354-4458,4564-4671,4878-5017 and the 5115-5281 of SEQ ID NO.5) be used to the sequence that is rich in T of the Transcription Termination that makes RNA polymerase 1 and/or 3, these introns are embedded in the cDTAwt encoding sequence; TGFP encoding sequence (nt6404-7095 of SEQ ID NO.5) under pSV40 promotor (nt.6044-6351 of the SEQ ID NO.5) control.This plasmid also comprises target site TD1 (SEQ ID NO.77), the TCTL (SEQ ID NO.80) of 8 copies and the TLacZ (SEQ ID NO.71) of 2 copies.
(6.E87 SEQ ID.NO.6)-Pcmv-4ORF
∧-TD1-3TLacZ-Tctl-BGlob-25G-XRN1S﹠amp; L-DT
∧A---Psv40-TGFP (pCMV promotor (nt.420-938 of SEQ ID NO.6); 4ORF
∧(nt.1027-3430 of SEQ ID NO.6); BGlob=adds beta Globulin 5 ' the brachymemma end (nt.3577-3655 of SEQ ID NO.6) of cap.25G=25 continuous G Nucleotide section (nt.3660-3684 of SEQ ID NO.6), described section can be hindered/disturb the XRN exoribonuclease; XRN1S﹠amp; L=can hinder stem and the ring structure (nt.3687-3767 of SEQ ID NO.6) of the yellow fever virus 3 ' UTR of XRN1 exoribonuclease.DT
∧A=contains from the intron of human collagen 16A1 gene and does not contain the kozak DTA (nt.3787-4711 of SEQ ID NO.6) of promotor/montage/poly a-signal; TGFP encoding sequence (nt6404-7095 of SEQ ID NO.6) under psv40 promotor (nt.5811-6502 of the SEQ ID NO.6) control.This plasmid also comprises TLacz (SEQ ID NO.71) and the TCTL target site (SEQ ID NO.80) of TD1 (SEQ ID NO.77), 3 copies.
(7.E123 SEQ ID.NO.7)-Psv40-intron-4ORF
∧-3X[TD1-TLacZ]-4PTE-SV40 intron-HBB-DTA---Phsvtk-Fluc (pSV40 promotor (nt.7-419 of SEQ ID NO.7), 4ORF
∧9 TISU sequences and 57 kozak sequences among=4 continuous ORF are 57,57,36,36,21,21,21 and 21nt (nt722-2387 of SEQ ID NO.7) between the adjacent ATG codon; 4 kinds of stems of 4PTE=palindrome termination element and ring structure (nt.3318-3473 of SEQ ID NO.7).The intron of the little t antigen of SV40 intron=SV40 (nt.3505-3596 of SEQ ID NO.7); HBB=does not contain ATG and comprises the oxyphorase β mRNA of its first intron (nt.3627-4406 of SEQ ID NO.7); CDTAwt encoding sequence (nt.4431-5014 of SEQ ID NO.7); HSKVK promotor (nt.5106-5858 of SEQ ID NO.7) and Lampyridea luciferase encoding sequence (nt.5894-7546 of SEQ ID NO.7).This plasmid also comprises TD1 (SEQ ID NO.77) and the TLacz target site (SEQ ID NO.71) of 3 copies.
(8.E30 SEQ ID.NO.8)-Pcmv-4ORF
∧-TD1-Tfluc-incDTAWT---Psv40-TGFP (pCMV promotor (nt.420-938 of SEQ ID NO.8); 4ORF
∧9 TISU sequences and 57 kozak sequences among=4 continuous ORF are 57,57,36,36,21,21,21 and 21nt (nt1027-3547 of SEQ ID NO.8) between the adjacent ATG codon.The one ORF (nt.1031-1651 of SEQ ID NO.8) is from TISU (nt.1027-1038 of SEQ ID NO.8) translation, and ensuing 3 ORF
∧(nt.1662-2996, nt.2306-2941 and the nt2951-3547 of SEQ ID NO.8) translates from the Kozak sequence.(cDTAwt=does not contain promotor/montage/termination/poly A site and contains the wt DTA encoding sequence of Kozak sequence (nt3568-4155 of SEQ ID NO.8) and stops before last ORF (nt2951-3516 of SEQ ID NO.8) at the encoding sequence of wild-type DTA; Next be the TGFP encoding sequence under the control of SV40 promotor.This plasmid also comprises target site TD1 (SEQ ID NO.77) and Tfluc (SEQ ID NO.74).
(9.E142 SEQ ID.NO.9)-3 poly A-Prpl19-4ORF
∧-TD1-Tfluc-S-cDTAWT---Phsvtk-Fluc.3 poly A=HSV poly A, SV40 poly A, synthetic poly A (nt.60-247 of SEQ ID NO.9); Prpl19=carries the promotor (nt.248-1941 of SEQ ID NO.9) of the RPL19 (ribosomal protein L 19) of its first intron; 4ORF
∧9 TISU sequences and 57 kozak sequences among=4 continuous ORF are 57,57,36,36,21,21,21 and 21nt (nt1948-4366 of SEQ ID NO.9) between the adjacent ATG codon; The encoding sequence of wild-type DTA (nt.4457-5044 of SEQ ID NO.9); The encoding sequence of HSKVK promotor (nt.5136-5888) and Lampyridea luciferase (nt.5924-7576 of SEQ ID.NO.9).This plasmid also comprises target site TD1 (SEQ ID NO.77) and Tfluc (SEQ ID NO.74).
The result:
Presented the result among following table 1-5 and the 6A-C.These results show under the various experiment conditions in transfection the RLU that measures in the cell of indicated plasmid and siRNA molecule.Used siRNA+ molecule is can be in conjunction with the siRNA molecule of its respective target sequence in tested plasmid.
Table 1:
Table 2:
Table 3:
Table 4:
Table 5:
Table 6A:
Table 6B:
Table 6C
About showing 6A-6C:
*=represent that described 2 kinds of siRNA+ show significant activation;
Plasmid E38 (the SEQ ID NO.19) cotransfection of *=also and 155ng.
More than the result who presents among table 1-5 and the 6A-6C shows in the presence of the siRNA molecule that can cause external source purpose RNA cracking, and exogenous object protein (DTA) is expressed, and it is expressed and causes necrocytosis to increase then.Described necrocytosis increase causes RLU observed value total in the hole to reduce, because cell expressing still less/generation luciferase genes.These results have confirmed, in fact, only in comprising the cell of specific siRNA, exogenous object protein (being DTA in the present embodiment) is expressed, because only in these cells, the cracking of external source purpose RNA at the cracking site place is initiated, thereby allows the expression of exogenous object protein in these cells.
Embodiment 2: composition of the present invention kills the purposes of relevant cancer of the stomach cancer cells, nasopharyngeal carcinoma cancer cells and Burkitt lymphoma cancer cells of EBV-.
Cancer of the stomach is the most general cancer and be the major cause of mortality ratio and sickness rate in the world after lung cancer.5 annual survival rates are less than 20%.The cancer of the stomach case in the whole world about 6 to 16% and the Ai Bositan epstein-Barr virus of in nearly all tumour, finding (EBV) relevant [21].Burkitt lymphoma is one type non-Hodgkin lymphoma, and it influences jawbone usually, forms big tumour group.The B cell is finally to cause the special lymphadenomatous the first step of Hugh Burkitt by the EBV immortalization.Nasopharyngeal carcinoma is the cancer of finding in the upper respiratory tract, and major part is usually at pharynx nasalis, and strong relevant with EBV virus.
Transplanting back lymphocytic hyperplasia illness (PTLPD) is the immunologic injury patient, for example suffers from the patient of AIDS or has experienced the another kind of B cell lymphoma that occurs among the patient of the organ transplantation relevant with immunosuppression, and therefore infer that it is relevant with EBV.The smooth muscle tumor of malignant tumor patient is also relevant with EBV with Hodgkin lymphoma.
In the U.S., the adult between 35 and 40 one full year of life of as many as 95% has infected Ai Bositan epstein-Barr virus (EBV or HHV-4).
The Ai Bositan epstein-Barr virus is coded in modulate tumor and suppresses 23 kinds of miRNA[13 of apoptosis aspect performance function].Two genome areas at the Ai Bositan epstein-Barr virus identify multiple miRNA and their expression [20] during the latent infection of the B clone of conversion.
At B cell Burkitt lymphoma, infected the expression [21] of observing EBV miRNA miR-BART1 (SEQ ID NO.41) in the nasopharyngeal carcinoma cell of the EBV cancer of the stomach (EBVaGC) relevant with EBV-.Therefore can kill these cancers by the composition of the present invention that the cell of expressing miR-BART1 is killed in use.
The ripe endogenous miRNA chain of EBV-mir-BART1 is: 5 '-UCUUAGUGGAAGUGACGUGCUGUG-3 ' (SEQ ID NO.42), the binding site of the exogenous RNA molecule of present embodiment is designed to comprise sequence: 3 '-AGAAUCACCUUCACUGCACGACAC-5 ' (SEQ ID NO.43), ripe endogenous miRNA chain 100% complementation of this sequence and EBV-mir-BART1.For example, referring to Figure 17.
The sequence of encoding exogenous target protein be designed to encode diphtheria toxin Segment A (DT-A) and be designed to be arranged in the downstream of exogenous RNA molecule EBV-mir-BART1 binding site.Be introduced in that unit molecule diphtheria toxin Segment A in the cell can be killed described cell [5] and in mammalian cell, the removal of cap makes the translation of mRNA reduce 35-50 doubly and makes the transformation period of functional mRNA reduce only 1.7 times [6].For example, referring to Figure 17.
Suppressing sequence is positioned at EBV-mir-BART1 binding site upstream and its and is designed to comprise and is positioned at people Kozak consensus sequence: 5 '-initiator codon of ACCAUGG-3 ' (SEQ ID NO.25) and be not in same reading frame with starting codon of DT-A.For example, referring to Figure 17.
The exogenous RNA molecule of present embodiment also comprises very effective cis acting set hammer head ribozyme-snorbozyme[15 at 5 ' end] before the exogenous RNA molecule is by the EBV-mir-BART1 cracking, to reduce the translation efficiency of described exogenous RNA molecule.Cis acting set hammer head ribozyme-snorbozyme also comprises 2 initiator codons, but its each be not in same reading frame with starting codon of DT-A.For example, referring to Figure 17.
The exogenous RNA molecule of present embodiment also comprises from the palindrome termination element (PTE) of 3 ' UTR of people HIST1H2AC (H2ac) gene in the sequence downstream of encoding D T-A (5 '-GGCUCUUUUCAGAGCC-3 '-SEQ ID NO.34).PTE mRNA processing and stable aspect play an important role [7].From HIST1H2AC gene transcription thing shortage poly (A) tail and owing to PTE is still stable.For example, referring to Figure 17.
In present embodiment shown in Figure 17, the exogenous RNA molecule is transcribed under the control of strong viral CMV promotor by virus vector.The sequence of the complete exogenous RNA molecule of present embodiment is listed with SEQ ID NO.44.
Transcribe described exogenous RNA molecule in the target cell of the carrier of the exogenous RNA molecule of having introduced code book embodiment after, cis acting type ribozyme is removed the cap of 5 ' end and to reduce any translation and the palindrome termination element of exogenous RNA molecule is stablized described exogenous RNA molecule and prevented that it is degraded.The initiator codon that does not meet reading frame stops the translation of DT-A, but when in target cell, having endogenous EBV-mir-BART1, the exogenous RNA molecule of present embodiment cleaved (sequence of cleaved sequence is listed with SEQ ID NO.45), and the initiator codon that does not meet reading frame is separated, make DT-A be translated and with the protein expression of at least one copy, the protein of described at least one copy is enough to cause necrocytosis.For example, referring to Figure 17.
Embodiment 3: composition of the present invention kills the purposes of HIV-1 cells infected
According to the World Health Organization, in 2006, the whole world 3,950 ten thousand people that have an appointment suffered from HIV.According to the estimation of United Nations about the project of HIV and AIDS, infer in Africa 9,000 ten thousand people's infected by HIV, causing minimum valuation is 1,800 ten thousand orphan.HIV (human immunodeficiency virus) can cause acquired immune deficiency syndrome (AIDS) (AIDS).Two class HIV infected person: HIV-1 and HIV-2.HIV-1 is more strong, relatively easily propagates and be the cause of disease of global most of HIV infection.HIV-2 propagates and mainly is confined to West Africa not too easily than HIV-1.
The many viruses that comprise HIV demonstrate resting stage or the latent period of carrying out few protein synthesis or not carrying out protein synthesis.During these stages, virus infection is sightless for immunity system basically.Present antiviral therapy scheme is invalid [1] in major part aspect the cell reserves of eliminating latent virus.
Immediate cause computer approach and high-throughput checking and the 109 kinds of Microrna precursors [13] by encoding viral of becoming feasible full genome screening and found.The nearest Microrna (for example miR-N367) that studies show that the HIV-1 coding is in influence and/or keep effect [1,14 and 19] in the latent infection.
In the human T-cell, the miRNA miR-N367 (SEQ ID NO.46) that expresses nef suppresses transcribe [19] of HIV-1.MiR-N367 by 5 '-LTR in the Negative Acknowledgment element in U3 zone reduce the activity [19] of HIV-1 LTR promotor.Therefore, the nef miRNA that produces in the cell of infected by HIV-1 can be by transcribing the back signal path and transcribing transcribe [19] that the neo-path is reduced HIV-1.
In present embodiment shown in Figure 180, composition of the present invention is designed to kill the cell that comprises endogenous miR-N367 (hiv1-mir-N367) and therefore also comprise HIV-1.
The ripe endogenous miRNA chain of miR-N367 is: 5 '-ACUGACCUUUGGAUGGUGCUUCAA-3 ' (SEQ ID NO.47), the binding site of the exogenous RNA molecule of present embodiment is designed to comprise the sequence with ripe miRNA chain 100% complementation of miR-N367: 5 '-UUGAAGCACCAUCCAAAGGUCAGU-3 ' (SEQ ID NO.48).(as shown in Figure 18).
The sequence of encoding exogenous target protein be designed to encode diphtheria toxin (DT) albumen and be designed to be arranged in the downstream of exogenous RNA molecule miR-N367 binding site.(Figure 18).
Suppress that sequence is positioned at miR-N367 binding site upstream and it is designed to comprise 2 initiator codons, one of them is positioned at people Kozak consensus sequence: 5 '-ACCAUGG-3 ' (SEQ ID NO.25) also its each be not in same reading frame with starting codon of DT.(Figure 18).
The exogenous RNA molecule also comprises the nucleotide sequence (SEQ ID NO.49) of 22 Nucleotide of the sequence upstream of miR-N367 binding site downstream and encoding D T albumen, wherein said nucleotide sequence can be in conjunction with the sequence (SEQ ID NO.50) of 22 Nucleotide in the sequence downstream that is positioned at encoding D T, wherein said exogenous RNA molecule forms the ring structure of the translation efficiency that increases DT, particularly when this exogenous RNA molecule is cleaved.
The exogenous RNA molecule also comprises very effective cis acting set hammer head ribozyme-N117[16 at 5 ' end] before the exogenous RNA molecule is by endogenous miRNA cracking, to reduce the translation efficiency of described exogenous RNA molecule.Cis acting set hammer head ribozyme-N117 also comprises 2 initiator codons, and it is not in same reading frame with starting codon of DT albumen.For example, referring to Figure 18.
In the present embodiment, the exogenous RNA molecule is transcribed under the control of strong viral CMV promotor by virus vector.The complete sequence of the exogenous RNA molecule of present embodiment is listed with SEQ ID NO.51.
Transcribe described exogenous RNA molecule in the target cell of the carrier of the exogenous RNA molecule of having introduced code book embodiment after, cis acting type ribozyme is removed the cap of 5 ' end to reduce any translation to described exogenous RNA molecule.The initiator codon that does not meet reading frame stops the translation of DT, but when in cell, having endogenous miR-N367 (or HIV-1), exogenous RNA molecule cleaved (sequence of cleaved sequence is listed with SEQ ID NO.52), and the initiator codon that does not meet reading frame is separated with the sequence of encoding D T albumen, makes DT to express.The RNA that comprises the sequence of encoding D T albumen partly forms the ring structure of increase DT protein translation to kill the cell that HIV-1 infects.For example, referring to Figure 18.
The virus vector of present embodiment also codified can strengthen the transcription factor (for example, NF-κ B) that the HIV1-miR-N367 in the cell that HIV-1 infects transcribes.This virus vector is the codified gene (Rev that for example, stops the HIV-1mRNA montage) that can stop new HIV-1 particle to produce also.
Embodiment 4: composition of the present invention kills the purposes of metastatic breast cancer cell
Compare with tumorigenicity cell healthy or that do not shift, in the metastatic breast cancer cell, [8] are raised in the expression of miR-10b (SEQ ID NO.53).The expression of miR-10b is raised [8] by transcription factor Twist.The target of miR-10b is the mobility [8] that the reduction of HOXD10 and HOXD10 level causes higher levels of RHOC and higher levels of RHOC irritation cancer cell.
In present embodiment shown in Figure 19, composition of the present invention is designed to kill the cell that comprises typical endogenous miR-10b in the metastatic breast cancer cell.
The ripe endogenous miRNA chain of miR-10b is: 5 '-UACCCUGUAGAACCGAAUUUGUG-3 ' (SEQ ID NO.54), the exogenous RNA molecule of present embodiment is designed to comprise 2 miR-10b binding sites, and in the wherein said binding site one comprises the sequence 5 '-CACAAAUUCGGUUCUACAGGGUA-3 ' (SEQ ID NO.55) [31] with ripe miRNA chain 100% complementation of miR-10b.(Figure 19).
The sequence of encoding exogenous target protein be designed to encode diphtheria toxin Segment A (DT-A) albumen and being designed between 2 miR-10b binding sites in the exogenous RNA molecule.In mammalian cell, the unit molecule diphtheria toxin Segment A that is introduced in the cell can be killed this cell [5].
The exogenous RNA molecule of present embodiment comprises 2 and suppresses sequences, and one at 5 ' end and another is at 3 ' end.
The inhibition sequence that is positioned at 5 of exogenous RNA molecule ' end is designed to comprise 3 initiator codons, one in the wherein said initiator codon is positioned at people Kozak consensus sequence: 5 '-ACCAUGG-3 ' (SEQ ID NO.25), and it is not in the same reading frame with starting codon of DT-A encoding sequence, and wherein all 3 initiator codons all are in the same reading frame.
The inhibition sequence that is positioned at 5 of exogenous RNA molecule ' end also comprises the nucleotide sequence of described 3 initiator codon downstreams and 2 miR-10b binding site upstreams, wherein said nucleotide sequence and described 3 initiator codons are in the same reading frame and wherein said nucleotide sequence coded sorting signals for Subcellular Localization, and described sorting signals is the peroxysome target signal 2 (H2N---RLRVLSGHL-SEQ ID NO.27) [28] of people's alkylphosphonic acid carboxylic acid dihydroxyacetone synthase.In mammalian cell, the protein that has for the sorting signals of Subcellular Localization can navigate to this subcellular location with its mRNA when it is translated.
The inhibition sequence that is positioned at 3 of exogenous RNA molecule ' end is designed to comprise the HSV1LAT intron in downstream, 2 miR-10b sites, wherein said exogenous RNA molecule is the target of decay (NMD) of the nonsense mediation of this exogenous RNA molecule of degraded, and the encoding sequence downstream of described exogenous RNA molecule in described exogenous RNA molecule comprises intron [29].
The inhibition sequence that is positioned at 3 of exogenous RNA molecule ' end also comprises the element that is rich in AU at 3 ' end place, and the described element that is rich in AU stimulates the degraded of exogenous RNA molecule.The length that is rich in the element of AU is that 47 Nucleotide and it comprise sequence: 5 '-AUUUA-3 ' (SEQ ID NO.31) and 5 '-UUAUUUA (U/A) is (U/A)-3 ' (SEQ ID NO.32) [26].
In the present embodiment, the exogenous RNA molecule is transcribed under the control of strong viral CMV promotor by virus vector.The sequence of the complete exogenous RNA molecule of present embodiment is listed with SEQ ID NO.56.
After in the target cell of the carrier of the exogenous RNA molecule of having introduced code book embodiment, transcribing described exogenous RNA molecule, the initiator codon that does not meet reading frame stops the translation of DT-A, peroxysome target signal 2 is sent to peroxysome with albumen and the exogenous RNA molecule of mistake, intron is molecular targeted decay (NMD) degraded for being mediated by nonsense of exogenous RNA, and the element that is rich in AU also stimulates the degraded of exogenous RNA molecule.Yet when in cell, having endogenous miR-10b, exogenous RNA molecule cleaved (sequence of cleaved sequence is listed with SEQ ID NO.57), and all inhibition sequences are separated, cause DT-A albumen to be translated and with the protein expression of at least one copy, the protein of described at least one copy is enough to cause necrocytosis.
Embodiment 5: composition of the present invention kills the purposes of HSV-1 cells infected
The many viruses that comprise HSV-1 (hsv-1) demonstrate resting stage or the latent period of carrying out few protein synthesis or not carrying out protein synthesis.During these stages, virus infection is sightless for immunity system basically.Present antiviral therapy scheme is invalid in major part aspect the cell reserves of eliminating latent virus.
The relevant transcript (LAT) of hiding of hsv-1 (HSV-1) is the virogene of unique expression during the neurone latent infection.LAT suppresses apoptosis and hides by impelling infected neuronal survival to keep.Do not belong to the protein of LAT gene.The miRNA-miR-LAT (SEQ ID NO.58) that studies show that HSV-1 LAT genes encoding provides the resistance to apoptosis [17].MiR-LAT produce from the exons 1 zone of HSV-1LAT gene and therefore miR-LAT during latent infection, express [17].
In present embodiment shown in Figure 20, composition of the present invention is designed to kill the cell that comprises endogenous miR-LAT and therefore also comprise HSV-1.
The ripe endogenous miRNA chain of miR-LAT is: 5 '-UGGCGGCCCGGCCCGGGGCC-3 ' (SEQ ID NO.59), and the exogenous RNA molecule of present embodiment is designed to comprise 2 miR-LAT binding sites, wherein each binding site comprise with the sequence 5 of ripe miRNA chain 100% complementation of miR-LAT '-GGCCCCGGGCCGGGCCGCCA-3 ' (SEQ ID NO.60) [17].
The sequence of encoding exogenous target protein be designed to encode diphtheria toxin (DT) albumen and be designed to be arranged in the downstream (Figure 20) of two miR-LAT binding sites of exogenous RNA molecule.
The exogenous RNA molecule comprises that also 2 are suppressed sequences, and one at 5 ' end and another is at 3 ' end.
The inhibition sequence that is positioned at 5 of exogenous RNA molecule ' end is designed to comprise 2 initiator codons, its each all be positioned at people Kozak consensus sequence: 5 '-ACCAUGG-3 ' (SEQ ID NO.25) and its be not in same reading frame (Figure 20) with starting codon of DT albumen.
The inhibition sequence that is positioned at 3 of exogenous RNA molecule ' end is designed to be included in the translation repressor smaug recognition component (SRE) in 2 miR-LAT binding site downstreams: 5 '-UGGAGCAGAGGCUCUGGCAGCUUUUGCAGCG-3 ' (SEQ ID NO.28).Smaug1 is coded in No. 14 karyomit(e)s of people and can checks the courier's who comprises SRE translation [24,25].Mouse Smaug1 is expressed in the brain and stimulated a closely subcellular area of regulation and control translation by cynapse, enriches in the synaptoneurosome.
The inhibition sequence that is positioned at 3 of exogenous RNA molecule ' end also comprises the RNA signal for locating that is used for the myelinization periphery at 3 ' end place (the ribonucleoprotein A2 response element of A2RE-nuclear): 5 '-GCCAAGGAGCCAGAGAGCAUG-3 ' (SEQ ID NO.29) [27].A2RE is to be positioned at the cis acting sequence of 3 of MBP (myelin basic protein) mRNA '-untranslated region and is enough and essential [27] for the myelinization periphery that MBP mRNA is delivered to oligodendrocyte.HnRNP (hnRNP) A2 is in conjunction with A2RE and mediate the transportation [27] of MBP.
The exogenous RNA molecule also comprises the tenuigenin polyadenylic acid element (CPE) in the sequence downstream of next-door neighbour's encoding D T albumen.CPE comprises the sequence 5 '-UUUUAUU-3 ' (SEQ ID NO.39) [23] at 91 Nucleotide places, sequence downstream of the sequence 5 '-UUUUUUAUU-3 ' (SEQ ID NO.38) in sequence downstream of next-door neighbour encoding D T albumen and encoding D T albumen.In Mammals, CPEB (tenuigenin polyadenylic acid element conjugated protein) is present in the dendritic layer [30] of hippocampus (brain is responsible for the zone of long-term memory).At cynapse-dendron compartment of Mammals hippocampal neuron, CPEB shows as the translation that stimulates the α-CaMKII mRNA that comprises CPE by the translation of polyadenylic acid initiation.
In the present embodiment, the exogenous RNA molecule is transcribed under the control of strong viral CMV promotor by virus vector.The sequence of the complete exogenous RNA molecule of present embodiment is listed with SEQ ID NO.61.
After in the target cell of the carrier of the exogenous RNA molecule of having introduced code book embodiment, transcribing described exogenous RNA molecule, the initiator codon that does not meet reading frame stops the translation of DT albumen, Smaug1 (translation repressor) in conjunction with smaug recognition component (SRE) and suppress the translation of DT albumen and hnRNP A2 in conjunction with A2RE and mediate the exogenous RNA molecule to the transportation of myelinization periphery.Yet when in target cell, having (HSV-1's) endogenous miR-LAT, exogenous RNA molecule cleaved (sequence of cleaved sequence is listed with SEQ ID NO.62), and 2 are suppressed sequence and separate, cause CPEB (tenuigenin polyadenylic acid element conjugated protein) in conjunction with CPE and stimulate the extension of the sweet sour tail of poly gland in the exogenous RNA molecule of cracking, make DT to express and therefore kill described cell and adjacent cell.
The reference tabulation
1.Weinberg M.S and Morris K.V, 2006.Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol25:223-231.
2.Velculescu people such as VE, 2006.The Consensus Coding Sequences of Human Breast and Colorectal Cancers, Science314,5797:268-274.
3.Lord MJ, Jolliffe NA, Marsden CJ waits the people, 2003, Ricin Mechanisms of Cytotoxicity, Toxicol Rev22,1:53-64.
4.Jeen-Kuan Chen, Chih-Hung Hung, Yen-Chywan Liaw and Jung-Yaw Lin, 1997, Identification of amino acid residues of Abrin-a A chain is essential for catalysis and reassociation with Abrin-a B chain by site-directed mutagenesis, Protein Engineering10:827-833.
5.Yamaizumi, M, Mekada, E, Uchida, T. and Okada Y, 1978, One molecule of Diphtheria toxin fragment A introduced into a cell can kill the cell, cell15,1:245-50.
6.Gallie?DR,1991,The?cap?and?poly(A)tail?function?synergistically?to?regulate?mRNA?translational?efficiency.Genes?Dev5,11:2108-16.
7.Entrez?Gene:HIST1H2AC?histone?cluster1,H2ac.
8.T.Dalmay,2008,MicroRNAs?and?cancer,Journal?of?Internal?Medicine263,4:366-375.
9.Y?Zeng,2006,Principles?of?micro-RNA?production?and?maturation,Oncogene25:6156-6162.
10.BM?Engels?and?G?Hutvagner,2006,Principles?and?effects?of?microRNA-mediated?post-transcriptional?gene?regulation,Oncogene?25:6163-6169.
11.Benjamin?Haley?&?Phillip?D?Zamore,2004,Kinetic?analysis?of?the?RNAi?enzyme?complex,Nature?Structural?&?Molecular?Biology11:599-606.
12.William?CS?Cho,2007,OncomiRs:the?discovery?and?progress?of?microRNAs?in?cancers.Molecular?Cancer6:60.
13.Vinod Scaria and Vaibhav Jadhav, 2007, microRNAs in viral oncogenesis, Retrovirology4:82.
14.Vinod?Scaria,Manoj?Hariharan,Beena?Pillai,Souvik?Maiti,Samir?K.Brahmachari,2007,Host-virus?genome?interactions:macro?roles?for?microRNAs,Cellular?Microbiology9,12:2784-2794.
15.Maurille people such as J.Fournier, 1999.A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency, PNAS96,12:6609-6614.
16.Laising?Yen,Jennifer?Svendsen,Jeng-Shin?Lee,John?T.Gray,Maxime?Magnier,Takashi?Baba,Robert?J.D′Amato?&?Richard?C.Mulligan,2004,Exogenous?control?of?mammalian?gene?expression?through?modulation?of?RNA?self-cleavage,Nature431,471-476.
17.Gupta,J.J.Gartner,P.Sethupathy,A.G.Hatzigeorgiou?&?N.W.Fraser,2006,Anti-apoptotic?function?of?a?microRNA?encoded?by?the?HSV-1?latency-associated?transcript,Nature442,82-85.
18.
http://microrna.sanger.ac.uk/cgi-bin/sequences/miRNA_entry.pl?acc=MI0000267
19.Shinya Omoto and Yoichi R.Fujii, 2005, Regulation of human immunodeficiency virus1transcription by nef microRNA, J Gen Virol86:751-755.
20.Grey?F,Meyers?H,White?EA,Spector?DH,Nelson?J,2007,A?Human?Cytomegalovirus-Encoded?microRNA?Regulates?Expression?of?Multiple?ViralGenes?Involved?in?Replication.PLoS?Pathog3,11:e163.doi:10.1371/journal.ppat.0030163.
21.Do Nyun Kim, Hiun-Suk Chae, Sang Taek Oh, Jin-Hyoung Kang, ChoHyun Park, Won Sang Park, Kenzo Takada, Jae Myun Lee, Won-Keun Lee, with Suk Kyeong Lee, 2007, Expression of Viral MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma, Journal of Virology 1033-1036 page or leaf, the 81st volume, the 2nd phase.
22.Chabanon Herve and Ian Mickleburgh, 2004, Zipcodes and postage stamps:mRNA localisation signals and their trans-acting binding proteins, Briefings in Functional Genomics and Proteomics3:240-256.
23.Wu?L,Wells?D,Tay?J,Mendis?D,Abbott?MA,Barnitt?A,Quinlan?E,Heynen?A,Fallon?JR,Richter?JD,1998,CPEB-mediated?cytoplasmic?polyadenylation?and?the?regulation?of?experience-dependent?translation?of?alpha-CaMKII?mRNA?at?synapses,Neuron21,5:936-8.
24.Maria V.Baez and Graciela L.Boccaccio, Career investigator of the ConsejoNacional de Investigaciones Cientificas y Tecnologicas, 2005, Mammalian Smaug Is a Translational Repressor That Forms Cytoplasmic Foci Similar to Stress Granules, J.Biol.Chem280,52:43131-43140.
25.C A Smibert, J E Wilson, K Kerr and P M Macdonald, 1996, smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo, Genes ﹠amp; Development10:2600-2609.
26.Carine Barreau, Luc Paillard and H.Beverley Osborne, 2006, AU-rich elements and associated factors:are there unifying principles? Nucleic Acids Research33,22:7138-7150.
27.Trent P.Munro, Rebecca J.Magee, Grahame J.Kidd, John H.Carson, Elisa Barbarese, Lisa M.Smith and Ross Smith, 1999, Mutational Analysis of a Heterogeneous Nuclear Ribonucleoprotein A2 Response Element for RNA Trafficking, J Biol Chem, 274,48:34389-34395.
28.Suresh?subramani,1998,Components?Involved?in?Peroxisome?Import,Biogenesis,Proliferation,Turnover,and?Movement,PHYSIOLOGICAL?REVIEWS78:171-188.
29.Isken?O,Maquat?LE,2007,Quality?control?of?eukaryotic?mRNA:safeguarding?cells?from?abnormal?mRNA?function,Genes?Dev21,15:1833-56.
30.Joel?D.Richter,2001,Think?globally,translate?locally:What?mitotic?spindles?and?neuronal?synapses?have?in?common,Proc?Natl?Acad?Sci?USA98,13:7069-7071.
31.http://microrna.sanger.ac.uk/cgi-bin/sequences/miRNA_entry.pl?acc=MI0000267
32.Michael S.Wollenberg and Sanford M.Simon, 2004, Signal Sequence Cleavage of Peptidyl-tRNA Prior to Release from the Ribosome and Translocon, J.Biol.Chem.279:24919-24922.
33.Dan Frumkin, Adam Wasserstrom, Shalev Itzkovitz, Tomer Stern, Alon Harmelin, Raya Eilam, Gideon Rechavi and Ehud Shapiro, 2008, Cell Lineage Analysis of a Mouse Tumor, Cancer Research68:5924-5931.
34.Ugo?Moens,2009,Silencing?Viral?MicroRNA?as?a?Novel?Antiviral?Therapy?Journal?of?Biomedicine?and?Biotechnology.
35.Zhumur Ghosh, Bibekanand Mallick and Jayprokas Chakrabarti, 2009, Cellular versus viral microRNAs in host-virus interaction.Nucleic Acids Res.
36.Michaelis?M,Doerr?HW,Cinatl?J.Neoplasia,2009,The?story?of?human?cytomegalovirus?and?cancer:increasing?evidence?and?open?questions.Neoplasia?Press.
37.Theodore?WH,Epstein?L,Gaillard?WD,Shinnar?S,Wainwright?MS,Jacobson?S,2008,Human?herpes?virus6B:a?possible?role?in?epilepsy?.Epilepsia.
38.Elfakess?R,Dikstein?R.(2008).A?translation?initiation?element?specific?to?mRNAs?with?very?short?5′UTR?that?also?regulates?transcription.PloS?One.2008Aug28;3(8):e3094.
Claims (42)
1. a composition comprises for instructing exogenous object protein one or more polynucleotide that only cell of miRNAs is expressed in expression specificity, and described one or more polynucleotide encodings comprise following exogenous RNA molecule:
A) sequence of the described exogenous object protein of coding;
B) can suppress the inhibition sequence that described exogenous object protein is expressed; With
C) at the binding site of miRNAs in the described specificity,
Wherein only in described specificity miRNAs in the presence of, described exogenous RNA molecule is cleaved at the cracking site place, thereby discharges described inhibition sequence from the sequence of the described exogenous object protein of described coding, makes described exogenous object protein to be expressed.
2. composition as claimed in claim 1, wherein said cracking site in described binding site and wherein said cracking site between the sequence of described inhibition sequence and the described exogenous object protein of described coding.
3. composition as claimed in claim 1, wherein said at miRNAs in the described specificity binding site and described specificity in sequence in the miRNAs have and make that miRNAs instructs described exogenous RNA molecule in enough complementarity of described cracking site place's cracking in the described specificity.
4. miRNAs is the Microrna of cell, viral Microrna or the two in the composition as claimed in claim 1, wherein said specificity.
5. composition as claimed in claim 1, wherein said endogenous Microrna is only expressed in neoplastic cell.
6. composition as claimed in claim 4, wherein said viral Microrna are by the expressing viral that is selected from by the following group of forming: double-stranded DNA virus, single-stranded DNA viruses, diplornavirus, diplornavirus, strand (normal chain) virus, strand (minus strand) virus and retrovirus.
7. composition as claimed in claim 1, wherein said exogenous object protein is toxin.
8. composition as claimed in claim 7, wherein said toxin is selected from the group of being made up of following: Ricin, ricin A chain, abrin, abrin A chain, diphtheria toxin A chain and modified forms thereof.
9. composition as claimed in claim 7, wherein said toxin is selected from the group of being made up of following: alpha toxin, saporin, Zea mays RIP, barley RIP, wheat RIP, corn RIP, rye RIP, flax RIP, shiga toxin, will are congratulated sample RIP, momordin, thymidine kinase, Pokeweed antiviral protein, are spent more white tree toxalbumin, pseudomonas (Pseudomonas) extracellular toxin, ETA, intestinal bacteria (Escherichia coli) Isocytosine deaminase and modified forms thereof.
10. composition as claimed in claim 1, wherein said inhibition sequence is positioned at described cracking site upstream, and wherein said inhibition sequence reduces the translation efficiency from the described exogenous object protein of described exogenous RNA molecule.
11. composition as claimed in claim 10, wherein said inhibition sequence comprises a plurality of initiator codons.
12. composition as claimed in claim 11, wherein the sequence of each described initiator codon and described encoding exogenous target protein is not in same reading frame.
13. composition as claimed in claim 11, wherein each described initiator codon mainly by 5 '-AUG-3 ' forms.
14. composition as claimed in claim 11, wherein each described initiator codon is positioned at the Kozak consensus sequence.
15. composition as claimed in claim 1, wherein said inhibition sequence can be in conjunction with polypeptide, and wherein said polypeptide reduces the translation efficiency from the described exogenous object protein of described exogenous RNA molecule.
16. composition as claimed in claim 15, wherein said polypeptide are translation repression albumen, wherein said translation repression albumen is endogenous translation repression albumen or by the translation repression albumen of described one or more polynucleotide encodings of described composition.
17. composition as claimed in claim 1, wherein said inhibition sequence comprise RNA signal for locating for Subcellular Localization, endogenous miRNA binding site or the two.
18. composition as claimed in claim 1, wherein said composition also comprise the polynucleotide sequence that coding can suppress the function RNA of endogenous exonuclease expression directly or indirectly.
19. composition as claimed in claim 1, wherein enough complementarity is at least 30% complementarity.
20. composition as claimed in claim 1, wherein enough complementarity is at least 90% complementarity.
21. composition as claimed in claim 1, wherein said binding site at miRNAs in the described specificity is a plurality of binding sites at identical or different endogenous miRNA, and wherein said cracking site is a plurality of cracking sites.
22. composition as claimed in claim 1, wherein said polynucleotide comprise one or more dna moleculars, one or more RNA molecules or its combination.
23. miRNAs is selected from the group of being made up of following: hsv1-miR-H1 in the composition as claimed in claim 1, wherein said specificity, hsv1-miR-H2, hsv1-miR-H3, hsv1-miR-H4, hsv1-miR-H5, hsv1-miR-H6, hsv2-miR-I, hcmv-miR-UL22A, hcmv-miR-UL36, hcmv-miR-UL70, hcmv-miR-UL112, hcmv-miR-UL148D, hcmv-miR-US4, hcmv-miR-US5-1, hcmv-miR-US5-2, hcmv-miR-US25-1, hcmv-miR-US25-2, hcmv-miR-US33, kshv-miR-K12-1, kshv-miR-K12-2, kshv-miR-K12-3, kshv-miR-K12-4, kshv-miR-K12-5, kshv-miR-K12-6, kshv-miR-K12-7, kshv-miR-K12-8, kshv-miR-K12-9, kshv-miR-K12-10a, kshv-miR-K12-10b, kshv-miR-K12-11, kshv-miR-K12-12, ebv-miR-BART1, ebv-miR-BART2, ebv-miR-BART3, ebv-miR-BART4, ebv-miR-BART5, ebv-miR-BART6, ebv-miR-BART7, ebv-miR-BART8, ebv-miR-BART9, ebv-miR-BART10, ebv-miR-BART11, ebv-miR-BART12, ebv-miR-BART13, ebv-miR-BART14, ebv-miR-BART15, ebv-miR-BART16, ebv-miR-BART17, ebv-miR-BART18, ebv-miR-BART19, ebv-miR-BART20, ebv-miR-BHRF1-1, ebv-miR-BHRF1-2, ebv-miR-BHRF1-3, bkv-miR-B1, jcv-miR-J1, hiv1-miR-H1, hiv1-miR-N367, hiv1-miR-TAR, sv40-miR-S1, MCPyV-miR-M1, hsv1-miR-LAT, hsv1-miR-LAT-ICP34.5, hsv2-miR-II, hsv2-miR-III, hcmv-miR-UL23, hcmv-miR-UL36-1, hcmv-miR-UL54-1, hcmv-miR-UL70-1, hcmv-miR-UL22A-1, hcmv-miR-UL112-1, hcmv-miR-UL148D-1, hcmv-miR-US4-1, hcmv-miR-US24, hcmv-miR-US33-1, hcmv-RNA β 2.7, ebv-miR-BART1-1, ebv-miR-BART1-2, ebv-miR-BART1-3, ebv-miR-BHFR1, ebv-miR-BHFR2, ebv-miR-BHFR3, hiv1-miR-TAR-5p, hiv1-miR-TAR-p, hiv1-HAAmiRNA, hiv1-VmiRNA1, hiv1-VmiRNA2, hiv1-VmiRNA3, hiv1-VmiRNA4, mir-675, hiv1-VmiRNA5, hiv2-miR-TAR2-5p, hiv2-miR-TAR2-3p, mdv1-miR-M1, mdv1-miR-M2, mdv1-miR-M3, mdv1-miR-M4, mdv1-miR-M5, mdv1-miR-M6, mdv1-miR-M7, mdv1-miR-M8, mdv1-miR-M9, mdv1-miR-M10, mdv1-miR-M11, mdv1-miR-M12, mdv1-miR-M13, mdv2-miR-M14, mdv2-miR-M15, mdv2-miR-M16, mdv2-miR-M17, mdv2-miR-M18, mdv2-miR-M19, mdv2-miR-M20, mdv2-miR-M21, mdv2-miR-M22, mdv2-miR-M23, mdv2-miR-M24, mdv2-miR-M25, mdv2-miR-M26, mdv2-miR-M27, mdv2-miR-M28, mdv2-miR-M29, mdv2-miR-M30, mcmv-miR-M23-1, mcmv-miR-M23-2, mcmv-miR-M44-1, mcmv-miR-M55-1, mcmv-miR-M87-1, mcmv-miR-M95-1, mcmv-miR-m01-1, mcmv-miR-m01-2, mcmv-miR-m01-3, mcmv-miR-m01-4, mcmv-miR-m21-1, mcmv-miR-m22-1, mcmv-miR-m59-1, mcmv-miR-m59-2, mcmv-miR-m88-1, mcmv-miR-m107-1, mcmv-miR-m108-1, mcmv-miR-m108-2, rlcv-miR-rL1-1, rlcv-miR-rL1-2, rlcv-miR-rL1-3, rlcv-miR-rL1-4, rlcv-miR-rL1-5, rlcv-miR-rL1-6, rlcv-miR-rL1-7, rlcv-miR-rL1-8, rlcv-miR-rL1-9, rlcv-miR-rL1-10, rlcv-miR-rL1-11, rlcv-miR-rL1-12, rlcv-miR-rL1-13, rlcv-miR-rL1-14, rlcv-miR-rL1-15, rlcv-miR-rL1-16, rrv-miR-rR1-1, rrv-miR-rR1-2, rrv-miR-rR1-3, rrv-miR-rR1-4, rrv-miR-rR1-5, rrv-miR-rR1-6, rrv-miR-rR1-7, mghv-miR-M1-1, mghv-miR-M1-2, mghv-miR-M1-3, mghv-miR-M1-4, mghv-miR-M1-5, mghv-miR-M1-6, mghv-miR-M1-7, mghv-miR-M1-8, mghv-miR-M1-9 and sv40-miR-S1.
24. composition as claimed in claim 1, wherein said exogenous RNA molecule also comprises the terminator codon between the starting codon of the sequence of described initiator codon and described coding target protein, and described termination codon and described initiator codon be in same reading frame, and wherein said terminator codon be selected from by 5 '-group that UAA-3 ', 5 '-UAG-3 ' and 5 '-UGA-3 ' forms.
25. composition as claimed in claim 1, wherein said inhibition sequence is positioned at the sequence upstream of the described exogenous object protein of described coding, wherein said inhibition sequence can form to have and be lower than-secondary structure of the folding free energy of 30kcal/mol, and wherein said secondary structure is enough to hinder the scanning rrna and arrives the described of described exogenous object protein and start codon.
26. composition as claimed in claim 1, wherein said cell is selected from the group of being made up of following: people's cell, zooblast, cultured cells and vegetable cell.
27. composition as claimed in claim 1, wherein said composition is introduced in the cell.
28. composition as claimed in claim 1, wherein said cell is present in the organism.
29. a diagnostic kit comprises the described composition of claim 1.
30. a pharmaceutical composition comprises the described composition of claim 1 and one or more vehicle.
31. one kind is used for the method that target kills target cell, described method comprises the described composition of claim 1 is incorporated in the described target cell that wherein said target cell comprises miRNAs in the described specificity.
32. a carrier comprises the polynucleotide sequence of encoding exogenous RNA molecule, wherein said exogenous rna molecule comprises:
A) sequence of encoding exogenous target protein;
B) can suppress the inhibition sequence that described exogenous object protein is expressed; With
C) at the binding site of miRNAs in the specificity.
33. carrier as claimed in claim 32, wherein said carrier is virus vector.
34. carrier as claimed in claim 32, wherein said carrier is non-virus carrier.
35. carrier as claimed in claim 32, wherein said at miRNAs in the described specificity binding site and specificity in sequence in the miRNAs have after described carrier being incorporated in the cell that comprises miRNAs in the described specificity, make that miRNAs instructs described exogenous RNA molecule in enough complementarity of described cracking site cracking in the described specificity.
36. carrier as claimed in claim 35, wherein said cracking site are positioned at described binding site at described specificity miRNAs, and wherein said cracking site is between the sequence of described inhibition sequence and the described exogenous object protein of described coding.
37. carrier as claimed in claim 36, miRNAs is the Microrna of cell, viral Microrna or the two in the wherein said specificity.
38. composition as claimed in claim 32, wherein said endogenous Microrna is only expressed in neoplastic cell.
39. composition as claimed in claim 37, wherein said viral Microrna are by the expressing viral that is selected from by the following group of forming: double-stranded DNA virus, single-stranded DNA viruses, diplornavirus, diplornavirus, strand (normal chain) virus, strand (minus strand) virus and retrovirus.
40. composition as claimed in claim 32, wherein said exogenous object protein is toxin.
41. composition as claimed in claim 40, wherein said toxin is selected from the group of being made up of following: Ricin, ricin A chain, abrin, abrin A chain, diphtheria toxin A chain and modified forms thereof.
42. composition as claimed in claim 40, wherein said toxin is selected from the group of being made up of following: alpha toxin, saporin, Zea mays RIP, barley RIP, wheat RIP, corn RIP, rye RIP, flax RIP, shiga toxin, will are congratulated sample RIP, momordin, thymidine kinase, Pokeweed antiviral protein, are spent more white tree toxalbumin, Pseudomonas exotoxin, ETA, coli cytosine deaminase and modified forms thereof.
Applications Claiming Priority (3)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| PCT/IL2010/000894 WO2012056440A1 (en) | 2010-10-28 | 2010-10-28 | COMPOSITIONS AND METHODS FOR ACTIVATING EXPRESSION BY A SPECIFIC ENDOGENOUS miRNA |
| ILPCT/IL2010/000894 | 2010-10-28 | ||
| PCT/IL2011/000837 WO2012056457A2 (en) | 2010-10-28 | 2011-10-27 | Compositions and methods for activating expression by a specific endogenous mirna |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| CN103314003A true CN103314003A (en) | 2013-09-18 |
Family
ID=45993239
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN2011800631535A Pending CN103314003A (en) | 2010-10-28 | 2011-10-27 | Compositions and methods for activating expression by specific endogenous miRNA |
Country Status (7)
| Country | Link |
|---|---|
| US (1) | US20130245096A1 (en) |
| EP (1) | EP2632932A4 (en) |
| JP (1) | JP2013544511A (en) |
| CN (1) | CN103314003A (en) |
| AU (1) | AU2011322114A1 (en) |
| CA (1) | CA2824604A1 (en) |
| WO (2) | WO2012056440A1 (en) |
Cited By (6)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN106574292A (en) * | 2014-01-10 | 2017-04-19 | 国立大学法人京都大学 | Method for identifying desired cell type using miRNA expression as indicator |
| CN108753836A (en) * | 2018-06-04 | 2018-11-06 | 北京大学 | A kind of gene regulation or editing system using RNA interference mechanisms |
| CN108841864A (en) * | 2018-06-04 | 2018-11-20 | 北京大学 | A kind of molecule sensor using RNA interference mechanism |
| CN110016501A (en) * | 2018-01-09 | 2019-07-16 | 江苏命码生物科技有限公司 | Marker, detection method and its application of unexplained fever |
| CN111057790A (en) * | 2019-12-11 | 2020-04-24 | 石河子大学 | Application of miRNA in preparation of kit for detecting KSHV latent infection |
| CN116716351A (en) * | 2023-03-30 | 2023-09-08 | 湖北天勤生物技术研究院有限公司 | Composition for constructing cynomolgus monkey Alzheimer's disease model, application and construction method thereof |
Families Citing this family (34)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP2056845B1 (en) | 2006-08-08 | 2017-10-11 | Rheinische Friedrich-Wilhelms-Universität Bonn | Structure and use of 5' phosphate oligonucleotides |
| US8524248B2 (en) | 2007-12-14 | 2013-09-03 | University of Pittsburgh—of the Commonwealth System of Higher Education | Methods to diagnose and immunize against the virus causing human Merkel cell carcinoma |
| WO2009141146A1 (en) | 2008-05-21 | 2009-11-26 | Gunther Hartmann | 5' triphosphate oligonucleotide with blunt end and uses thereof |
| EP2508530A1 (en) | 2011-03-28 | 2012-10-10 | Rheinische Friedrich-Wilhelms-Universität Bonn | Purification of triphosphorylated oligonucleotides using capture tags |
| EP2766498B1 (en) | 2011-10-14 | 2019-06-19 | President and Fellows of Harvard College | Sequencing by structure assembly |
| US11021737B2 (en) | 2011-12-22 | 2021-06-01 | President And Fellows Of Harvard College | Compositions and methods for analyte detection |
| WO2013096851A1 (en) | 2011-12-22 | 2013-06-27 | President And Fellows Of Harvard College | Compositions and methods for analyte detection |
| WO2013184754A2 (en) | 2012-06-05 | 2013-12-12 | President And Fellows Of Harvard College | Spatial sequencing of nucleic acids using dna origami probes |
| US10822420B2 (en) * | 2012-09-13 | 2020-11-03 | Chugai Seiyaku Kabushiki Kaisha | Gene knock-in non-human animal |
| EP2712870A1 (en) | 2012-09-27 | 2014-04-02 | Rheinische Friedrich-Wilhelms-Universität Bonn | Novel RIG-I ligands and methods for producing them |
| US10138509B2 (en) | 2013-03-12 | 2018-11-27 | President And Fellows Of Harvard College | Method for generating a three-dimensional nucleic acid containing matrix |
| CN104419749B (en) * | 2013-08-22 | 2017-02-15 | 江苏命码生物科技有限公司 | MicroRNA used for predicting curative effect of interferon on chronic HBV and application thereof |
| WO2016007839A1 (en) * | 2014-07-11 | 2016-01-14 | President And Fellows Of Harvard College | Methods for high-throughput labelling and detection of biological features in situ using microscopy |
| WO2016100812A1 (en) * | 2014-12-19 | 2016-06-23 | Moderna Therapeutics, Inc. | Terminal modifications of polynucleotides |
| CN107708710A (en) | 2015-03-17 | 2018-02-16 | 嵌合体生物工程公司 | Smart CAR devices, DE CAR polypeptides, Side CAR and its use |
| WO2017062513A1 (en) * | 2015-10-05 | 2017-04-13 | Modernatx, Inc. | Methods for therapeutic administration of messenger ribonucleic acid drugs |
| CA3004285A1 (en) | 2015-11-03 | 2017-05-11 | President And Fellows Of Harvard College | Method and apparatus for volumetric imaging of a three-dimensional nucleic acid containing matrix |
| US11052111B2 (en) | 2015-12-08 | 2021-07-06 | Chimera Bioengineering, Inc. | Smart CAR devices and DE CAR polypeptides for treating disease and methods for enhancing immune responses |
| WO2017189525A1 (en) | 2016-04-25 | 2017-11-02 | President And Fellows Of Harvard College | Hybridization chain reaction methods for in situ molecular detection |
| WO2017201019A1 (en) | 2016-05-17 | 2017-11-23 | Chimera Bioengineering, Inc. | Methods for making novel antigen binding domains |
| KR20190053180A (en) * | 2016-07-26 | 2019-05-17 | 센티 바이오사이언시스, 인코포레이티드 | Spatio-temporal Adjusters |
| CN109983125A (en) | 2016-08-31 | 2019-07-05 | 哈佛学院董事及会员团体 | The method for generating the nucleic acid sequence library for detecting by fluorescent in situ sequencing |
| GB2569252A (en) | 2016-08-31 | 2019-06-12 | Harvard College | Methods of combining the detection of biomolecules into a single assay using fluorescent in situ sequencing |
| JP7045378B2 (en) | 2016-09-01 | 2022-03-31 | キメラ・バイオエンジニアリング,インコーポレーテッド | Gold-optimized CAR T cells |
| DE102017103383A1 (en) * | 2017-02-20 | 2018-08-23 | aReNA-Bio GbR (vertretungsberechtigter Gesellschafter: Dr. Heribert Bohlen, 50733 Köln) | System and method for cell-type specific translation of RNA molecules in eukaryotes |
| CN109789167A (en) | 2017-04-14 | 2019-05-21 | 哈佛学院董事及会员团体 | Method for generating cell-derived microfilament network |
| WO2019027869A1 (en) * | 2017-07-31 | 2019-02-07 | Massachusetts Institute Of Technology | Rna cleavage-induced transcript stabilizer and uses thereof |
| WO2019160815A1 (en) | 2018-02-13 | 2019-08-22 | Chimera Bioengineering, Inc. | Coordinating gene expression using rna destabilizing elements |
| CN112770776A (en) | 2018-07-30 | 2021-05-07 | 瑞德库尔有限责任公司 | Method and system for sample processing or analysis |
| CN109628489B (en) * | 2019-01-07 | 2022-09-23 | 新乡医学院 | Method for improving expression level of CHO cell recombinant protein and application thereof, expression vector, expression system and preparation method thereof |
| US20210046117A1 (en) | 2019-08-18 | 2021-02-18 | Chimera Bioengineering, Inc. | Combination Therapy with Gold Controlled Transgenes |
| WO2022182697A1 (en) * | 2021-02-23 | 2022-09-01 | Board Of Regents, The University Of Texas System | A novel rna-based approach to cancer treatment |
| EP4355882A2 (en) * | 2021-06-15 | 2024-04-24 | Modernatx, Inc. | Engineered polynucleotides for cell-type or microenvironment-specific expression |
| WO2023100955A1 (en) * | 2021-11-30 | 2023-06-08 | 国立大学法人京都大学 | Rna molecule |
Citations (6)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN1615369A (en) * | 2001-11-29 | 2005-05-11 | 富卡斯吉诺米克斯有限责任公司 | Method for analyzing translation-controlled gene expression |
| WO2006073727A2 (en) * | 2004-12-21 | 2006-07-13 | Monsanto Technology, Llc | Recombinant dna constructs and methods for controlling gene expression |
| US20060265771A1 (en) * | 2005-05-17 | 2006-11-23 | Lewis David L | Monitoring microrna expression and function |
| US20090286242A1 (en) * | 2007-12-10 | 2009-11-19 | Cold Spring Harbor Laboratory | MicroRNA Expression Profiling and Uses Thereof |
| WO2009142602A1 (en) * | 2008-05-19 | 2009-11-26 | Agency For Science, Technology And Research | Nucleic acid molecule and method of targeting gene expression to gliomas |
| WO2010055413A1 (en) * | 2008-11-12 | 2010-05-20 | Fondazione Centro San Raffaele Del Monte Tabor | Gene vector for inducing transgene-specific immune tolerance |
Family Cites Families (6)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CA2405499A1 (en) * | 2000-03-28 | 2001-10-04 | Maurice Zauderer | Methods of producing a library and methods of selecting polynucletides |
| US20060275262A1 (en) * | 2001-07-26 | 2006-12-07 | Mathis James M | Conditionally replicating viruses and methods for cancer virotherapy |
| US20070265220A1 (en) * | 2004-03-15 | 2007-11-15 | City Of Hope | Methods and compositions for the specific inhibition of gene expression by double-stranded RNA |
| US20070179113A1 (en) * | 2005-05-19 | 2007-08-02 | Schering Aktiengesellachaft | GM-CSF gene therapy for Crohn's disease using an improved regulated expression system |
| US20070054872A1 (en) * | 2005-08-24 | 2007-03-08 | Mirus Bio Corporation | Regulatable or conditional expression systems |
| US20090156535A1 (en) * | 2007-09-27 | 2009-06-18 | The Trustees Of Princeton University | MicroRNAs for Modulating Herpes Virus Gene Expression |
-
2010
- 2010-10-28 WO PCT/IL2010/000894 patent/WO2012056440A1/en active Application Filing
-
2011
- 2011-10-27 WO PCT/IL2011/000837 patent/WO2012056457A2/en active Application Filing
- 2011-10-27 JP JP2013535589A patent/JP2013544511A/en active Pending
- 2011-10-27 EP EP11835739.1A patent/EP2632932A4/en not_active Withdrawn
- 2011-10-27 US US13/881,350 patent/US20130245096A1/en not_active Abandoned
- 2011-10-27 CN CN2011800631535A patent/CN103314003A/en active Pending
- 2011-10-27 AU AU2011322114A patent/AU2011322114A1/en not_active Abandoned
- 2011-10-27 CA CA2824604A patent/CA2824604A1/en not_active Abandoned
Patent Citations (6)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN1615369A (en) * | 2001-11-29 | 2005-05-11 | 富卡斯吉诺米克斯有限责任公司 | Method for analyzing translation-controlled gene expression |
| WO2006073727A2 (en) * | 2004-12-21 | 2006-07-13 | Monsanto Technology, Llc | Recombinant dna constructs and methods for controlling gene expression |
| US20060265771A1 (en) * | 2005-05-17 | 2006-11-23 | Lewis David L | Monitoring microrna expression and function |
| US20090286242A1 (en) * | 2007-12-10 | 2009-11-19 | Cold Spring Harbor Laboratory | MicroRNA Expression Profiling and Uses Thereof |
| WO2009142602A1 (en) * | 2008-05-19 | 2009-11-26 | Agency For Science, Technology And Research | Nucleic acid molecule and method of targeting gene expression to gliomas |
| WO2010055413A1 (en) * | 2008-11-12 | 2010-05-20 | Fondazione Centro San Raffaele Del Monte Tabor | Gene vector for inducing transgene-specific immune tolerance |
Non-Patent Citations (1)
| Title |
|---|
| BRIAN D BROWN,等: ""Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer"", 《NATURE MEDICINE》, vol. 12, no. 5, 23 April 2006 (2006-04-23), pages 585 - 591, XP002652777, DOI: 10.1038/nm1398 * |
Cited By (10)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN106574292A (en) * | 2014-01-10 | 2017-04-19 | 国立大学法人京都大学 | Method for identifying desired cell type using miRNA expression as indicator |
| CN110016501A (en) * | 2018-01-09 | 2019-07-16 | 江苏命码生物科技有限公司 | Marker, detection method and its application of unexplained fever |
| CN108753836A (en) * | 2018-06-04 | 2018-11-06 | 北京大学 | A kind of gene regulation or editing system using RNA interference mechanisms |
| CN108841864A (en) * | 2018-06-04 | 2018-11-20 | 北京大学 | A kind of molecule sensor using RNA interference mechanism |
| CN108753836B (en) * | 2018-06-04 | 2021-10-12 | 北京大学 | Gene regulation or editing system utilizing RNA interference mechanism |
| CN108841864B (en) * | 2018-06-04 | 2021-10-15 | 北京大学 | Molecular sensor using RNA interference mechanism |
| CN111057790A (en) * | 2019-12-11 | 2020-04-24 | 石河子大学 | Application of miRNA in preparation of kit for detecting KSHV latent infection |
| CN111057790B (en) * | 2019-12-11 | 2022-08-30 | 石河子大学 | Application of miRNA in preparation of kit for detecting KSHV latent infection |
| CN116716351A (en) * | 2023-03-30 | 2023-09-08 | 湖北天勤生物技术研究院有限公司 | Composition for constructing cynomolgus monkey Alzheimer's disease model, application and construction method thereof |
| CN116716351B (en) * | 2023-03-30 | 2024-02-23 | 湖北天勤生物技术研究院有限公司 | Composition for constructing cynomolgus monkey Alzheimer's disease model, application and construction method thereof |
Also Published As
| Publication number | Publication date |
|---|---|
| WO2012056457A2 (en) | 2012-05-03 |
| EP2632932A2 (en) | 2013-09-04 |
| EP2632932A4 (en) | 2014-12-17 |
| CA2824604A1 (en) | 2012-05-03 |
| WO2012056457A3 (en) | 2012-08-02 |
| JP2013544511A (en) | 2013-12-19 |
| AU2011322114A1 (en) | 2013-05-30 |
| US20130245096A1 (en) | 2013-09-19 |
| WO2012056440A1 (en) | 2012-05-03 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| CN103314003A (en) | Compositions and methods for activating expression by specific endogenous miRNA | |
| JP7306696B2 (en) | RNA-guided nucleic acid-modifying enzyme and method of use thereof | |
| JP2023027277A (en) | Rna-guided nucleic acid modifying enzymes and methods of use thereof | |
| JP7402163B2 (en) | Materials and methods for the treatment of Usher syndrome type 2A | |
| KR20210053898A (en) | New CRISPR enzyme and system | |
| JP2021503945A (en) | Materials and Methods for the Treatment of Autosomal Dominant Retinitis Pigmentosa | |
| CN109843914A (en) | For treating the material and method of pain associated disorder | |
| KR102621539B1 (en) | Engineered SC function-controlling system | |
| US11760983B2 (en) | Enhanced hAT family transposon-mediated gene transfer and associated compositions, systems, and methods | |
| US10995328B2 (en) | Materials and methods for treatment of autosomal dominant cone-rod dystrophy | |
| CN107405357A (en) | Multiple shRNAs and its application | |
| CN103282372A (en) | Compositions and methods for specific cleavage of exogenous rna in a cell | |
| CN109415729A (en) | With the gene editing reagent for reducing toxicity | |
| JP7305534B2 (en) | Materials and methods for treating pain-related disorders | |
| JP2022522650A (en) | CRISPR-CAS effector polypeptide and how to use it | |
| Arango-Lievano et al. | Cell-type specific expression of p11 controls cocaine reward | |
| US20090023670A1 (en) | Regulation of Transgene Expression by RNA Interference | |
| CN108866058B (en) | KRAS-targeted siRNA and application thereof in preparation of pancreatic cancer treatment drug | |
| WO2019113061A1 (en) | Compositions and methods for modulating gene expression | |
| US20240117350A1 (en) | Use of mirna-485 inhibitor to regulate psd95, synaptophysin, and caspase-3 expression | |
| EA045278B1 (en) | RNA-GUIDED NUCLEIC ACIDS MODIFYING ENZYMES AND METHODS OF THEIR APPLICATION | |
| WO2013018096A1 (en) | Use of integrase for targeted gene expression |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| C06 | Publication | ||
| PB01 | Publication | ||
| C10 | Entry into substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| C02 | Deemed withdrawal of patent application after publication (patent law 2001) | ||
| WD01 | Invention patent application deemed withdrawn after publication |
Application publication date: 20130918 |